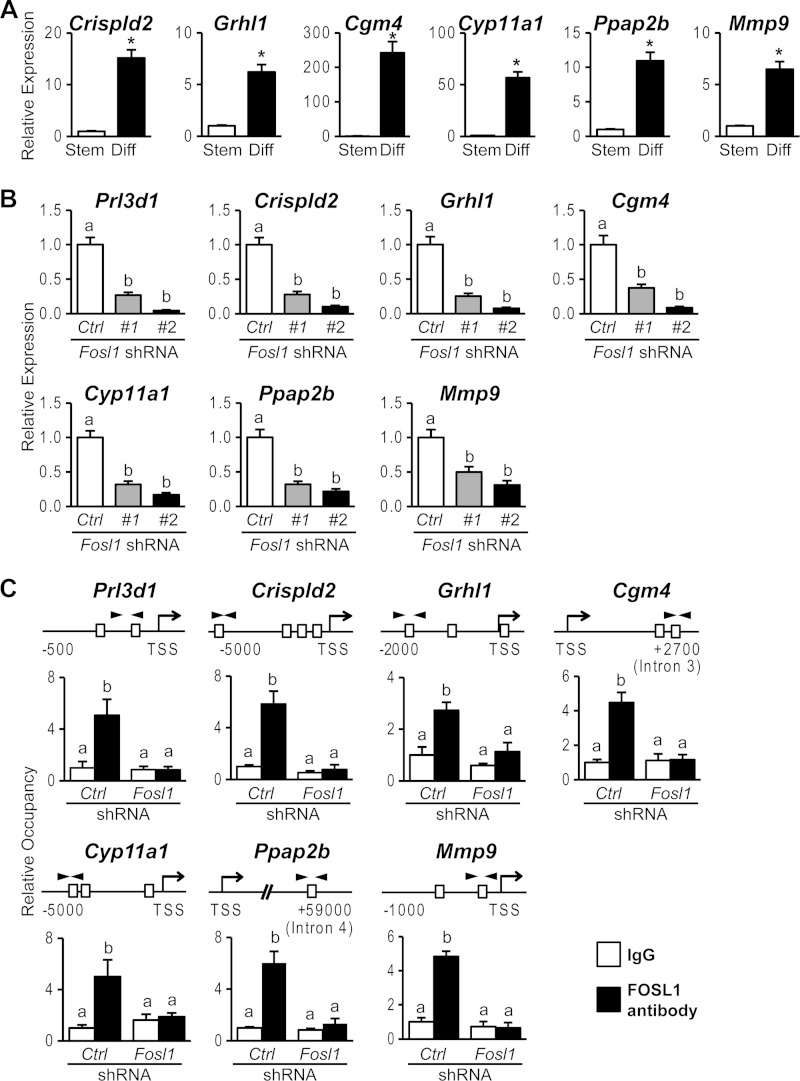
FIG 2.
FOSL1-dependent genes activated in trophoblast cells differentiating in vitro. (A) qRT-PCR analyses of Crispld2, Grhl1, Cgm4, Cyp11a1, Ppap2b, and Mmp9 transcripts in stem (Stem) or differentiated (Diff) Rcho-1 TS cells. (B) qRT-PCR analyses of Prl3d1, Crispld2, Grhl1, Cgm4, Cyp11a1, Ppap2b, and Mmp9 transcripts in differentiated Rcho-1 TS cells expressing control (Ctrl) or Fosl1 shRNAs. (C) FOSL1 occupancy at DNA regions possessing putative AP-1 binding elements within Prl3d1, Crispld2, Grhl1, Cgm4, Cyp11a1, Ppap2b, and Mmp9 loci. ChIP was used for determination of FOSL1 occupancy in differentiated Rcho-1 TS cells expressing control or Fosl1 shRNAs. Schematic representations of putative AP-1 elements (boxes) at each gene locus and the location of primers used to amplify regions containing the putative AP-1 elements (arrowheads) are shown at the top. IgG was included as a negative control. Bars represent means ± SEM. Values with an asterisk or different characters indicate significant differences (P < 0.05). TSS, transcription start site.