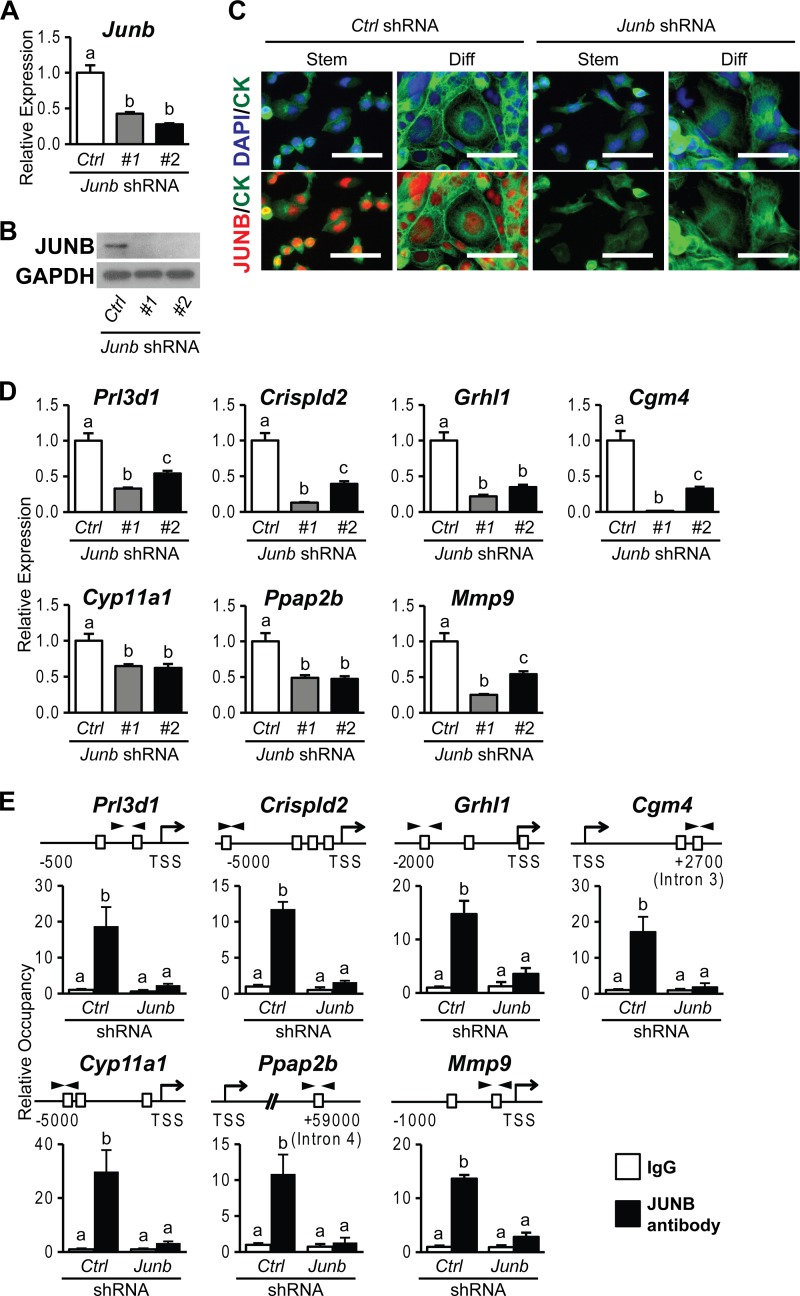
FIG 7.
JUNB-dependent genes activated in trophoblast cells differentiating in vitro. (A and B) JUNB knockdown efficiency in differentiated Rcho-1 TS cells expressing control (Ctrl) or Junb shRNAs was validated by qRT-PCR (A) and Western blot (B) analyses. GAPDH was used as an internal control. (C) JUNB localization and knockdown efficiency in stem (Stem) and differentiated (Diff) Rcho-1 TS cells expressing control or Junb shRNAs were assessed by immunocytochemistry. Cells were immunostained for JUNB and counterstained with cytokeratin (CK) and/or DAPI. Bar = 100 μm. (D) qRT-PCR analysis of Prl3d1, Crispld2, Grhl1, Cgm4, Cyp11a1, Ppap2b, and Mmp9 transcripts in differentiated Rcho-1 TS cells expressing control or Junb shRNAs. (E) ChIP analysis of JUNB occupancy in differentiated Rcho-1 TS cells expressing control or Junb shRNAs. Schematic representations of putative AP-1 elements (boxes) at each gene locus and the location of primers used to amplify regions containing the putative AP-1 elements (arrowheads) are shown at the top. IgG was included as a negative control. Bars in the histograms represent means ± SEM. Different letters above each bar indicate significant differences (P < 0.05).