Abstract
ADP-ribosyltransferase diphtheria-toxin like 1/poly(ADP-ribose) polymerase 1 (ARTD1/PARP1) is a chromatin-associated protein in the nucleus and plays an important role in different cellular processes such as regulation of gene transcription. ARTD1 has been shown to coregulate the inflammatory response by modulating the activity of the transcription factor nuclear factor κB (NF-κB), the principal regulator of interleukin 6 (IL-6), an important inflammatory cytokine implicated in a variety of diseases such as cancer. However, to what extent and how ARTD1 regulates IL-6 transcription has not been clear. Here, we show that ARTD1 suppresses lipopolysaccharide (LPS)-induced IL-6 expression in macrophages, without affecting the recruitment of the NF-κB subunit RelA to the IL-6 promoter and independent of its enzymatic activity. Interestingly, knockdown of ARTD1 did not alter H3 occupancy but increased LPS-induced trimethylation of histone 3 at lysine 4 (H3K4me3), a hallmark of transcriptionally active genes. We found that ARTD1 mediates its effect through the methyltransferase MLL1, by catalyzing H3K4me3 at the IL-6 promoter and forming a complex with NF-κB. These results demonstrate that ARTD1 modulates IL-6 expression by regulating the function of an NF-κB enhanceosome complex, which involves MLL1 and does not require ADP-ribosylation.
INTRODUCTION
Interleukin 6 (IL-6) is an important inflammatory cytokine triggered, e.g., by pathogen-associated molecular patterns (PAMPs) such as bacterial lipopolysaccharide (LPS) (1). Increased IL-6 production is linked to inflammatory diseases such as inflammatory bowel disease, rheumatoid arthritis, mast cell growth proliferation, chronic inflammation, and obesity and to different cancers such as breast, colon, epithelial, or lung cancer (1–6). IL-6 can be released from tumors themselves, or from cancer-associated fibroblasts, and together with other factors thereby creates a tumor-promoting microenvironment (7–9). Understanding the mechanisms how IL-6 transcription is regulated in different cell types is thus important for diseases such as cancer and inflammatory diseases. While the expression of IL-6 is principally regulated by the transcription factor nuclear factor κB (NF-κB), epigenetic mechanisms also play an important role in the regulation of IL-6 gene expression (10–14).
NF-κB is a widely expressed, inducible transcription factor crucial for inflammation, immunity, cell proliferation, and apoptosis (15, 16). In mammalian cells, five members of the NF-κB family exist, forming different homo- or heterodimers (17). The most abundant, best-studied, and “classical” form of NF-κB is a heterodimer consisting of the two subunits p50 and RelA (p65). In unstimulated cells, NF-κB is mostly sequestered in the cytoplasm as an inactive transcription factor complex by its physical association with one of the several inhibitors of NF-κB (IκB). PAMPs induce the classical, canonical pathway, which involves the rapid activation of IκB kinase β (IKKβ), NEMO-dependent phosphorylation and the subsequent degradation of IκBs, and the consequent nuclear translocation of primarily RelA-containing NF-κB heterodimers. As a mechanism to control the inflammatory response, nuclear NF-κB activity is selectively regulated at various levels downstream of activation, including DNA methylation (which depends on the differentiation state of the cell in question), nucleosome positioning, and histone modifications (e.g., histone methylation such as H3K4, H3K4, or H4K27 methylation), and via complex formation with coregulators, including p300/CBP and MLL1 (18–22). MLL1 is a member of the SET1/MLL family of methyltransferases and is known for its crucial functions for homeobox gene expression during development and embryogenesis and for stem cell regulation, as well as for gene transcription in general (23, 24). Interestingly, MLL1-dependent regulation of NF-κB downstream genes, including IL-6, has recently been reported (25). Due to these pivotal functions, mutations affecting the MLL1 gene cause severe diseases and are implicated in acute leukemia in children and adults, with a particularly poor prognosis (26). To prevent fatal malfunction or misregulation of MLL1, multiple mechanisms control its activity in the cell (27). Together, there is a diversity of regulatory mechanisms for the differential activation of NF-κB-dependent target genes within the same cell or the differential activation of the same gene in different cells (20, 28). Furthermore, NF-κB is subject to positive-feedback regulation by cytokines such as IL-6 (29, 30).
ADP-ribosyltransferase diphtheria-toxin like 1/poly(ADP-ribose) polymerase 1 (ARTD1/PARP1) is an abundant nuclear protein that plays key roles in a variety of nuclear processes, including the regulation of transcription (31). ARTD1 possesses an intrinsic enzymatic activity that catalyzes the transfer of ADP-ribose (ADPr) units from NAD (NAD+) onto target gene regulatory proteins, thereby modulating their activities, functions, and interacting partners (32, 33).
Since its discovery, most studies on ARTD1 have focused on its role in DNA damage detection and repair responses (34). However, over the past decade, the role of ARTD1 in gene regulation has received increasing attention (31). Interestingly, ARTD1 can act as a transcriptional enhancer or as an attenuator. ARTD1 can regulate transcription by binding to nucleosomes and interacts dynamically with different types of chromatin domains to modulate the chromatin structure (34, 35). Nucleosome binding and auto-ADP-ribosylation of ARTD1 has been described as the underlying mechanism for this formation of transcriptionally inactive, dense chromatin (36, 37) and have been implicated in the reciprocal binding of ARTD1 and histone H1 to chromatin (38). ARTD1 has also been shown to covalently modify histone and chromatin-associated nonhistone proteins with poly(ADP-ribose) (PAR) (39, 40). ARTD1 can modulate the activity of nucleosome remodelers through noncovalent mechanisms, as is the case with ALC1 (amplified in liver cancer 1; also known as CHD1L), a macrodomain-containing nucleosome-remodeling enzyme. PAR-dependent interactions between ARTD1 and ALC1 promote nucleosome remodeling by ALC1, as well as recruitment of ALC1 to sites of DNA damage in cells (41, 42). ADP-ribosylation of KDM5B, a histone lysine demethylase acting on H3 lysine 4 trimethylation (H3K4me3), has been shown to block the binding of KDM5B to chromatin and inhibit its demethylase activity (43). This antagonism between ARTD1 and KDM5B helps to explain the high correlation between ARTD1 and H3K4me3 at actively transcribed promoters. The functional interplay between ARTD1 and KDM5B helps to control the chromatin state at ARTD1-regulated promoters for both basal and signal-regulated transcriptional outcomes (43). Finally, ARTD1 may also function as a scaffold protein independent of its catalytic activities, by interacting with and promoting the recruitment of other coregulatory enzymes required for transcription. We were the first to show that ARTD1 directly interacts with the NF-κB subunits and thereby regulates NF-κB-dependent gene expression (44, 45). We found that ARTD1 synergistically coregulates transcription together with known NF-κB transcriptional cofactors such as p300, CARM1, PRMT1 and the Mediator complex (18, 45, 46).
Although the enzymatic activity is not required for the transcriptional activation of transiently transfected NF-κB reporter plasmids by ARTD1 or upon NLRP3 inflammasome-induced ARTD1 cleavage (47), we were able to link ARTD1 and ADP-ribosylation to signaling during inflammation and the expression of adhesion molecules in atherogenesis, as well as cell survival under stress conditions (48). ARTD1 and NF-κB are thus interconnected in the inflammatory response.
At the inflammatory level, we have recently shown that nonapoptotic LPS-induced caspase 7 activation via the NLRP3 inflammasome induces ARTD1 cleavage at the transcriptional start sites (TSS) of distinct NF-κB target genes, including IL-6, and thereby causes elevated expression of these genes (49). The molecular mechanism responsible for the repressed IL-6 expression levels in the presence of ARTD1 was not elucidated, however.
In the present work, we characterized and elucidated the molecular mechanism by which ARTD1 regulates the transcription of IL-6. Our results demonstrate that the negative regulation of IL-6 expression by ARTD1 is independent of its enzymatic activity and does not affect RelA recruitment to the IL-6 promoter. Instead, we found that ARTD1 is enriched at the IL-6 promoter and suppresses MLL1-dependent H3K4me3. These results uncover a new mechanism of chromatin remodeling by ARTD1 and its importance for inflammation.
MATERIALS AND METHODS
Cell culture.
Mouse leukemic monocyte macrophage cell line (RAW 264.7) was cultured in RPMI medium (Gibco/Invitrogen, Carlsbad, CA) at 37°C. NIH 3T3 and HEK293T cells were cultivated in Dulbecco modified Eagle medium (PAA, Pasching, Austria). Bone marrow-derived macrophages (BMDM) were obtained from bones (femurs and tibias) of wild-type and ARTD1 knockout mice and also cultivated in RPMI medium. The bones were cut from both ends, and bone marrow was flushed out with complete medium using a 1-ml syringe with a 23G needle until the bones were completely white. Cells were resuspended in RPMI medium supplemented with 20% of the supernatant of L929 cells and plated in bacterial dishes for 5 days. Differentiated macrophages were maintained in culture for 2 weeks in RPMI medium supplemented with 5% L929 supernatant. All media were supplemented with 1% (vol/vol) penicillin-streptavidin and 10% (vol/vol) fetal calf serum (Gibco/Invitrogen).
Cells were preincubated with the ARTD1 inhibitors olaparib (1 μM; SelleckChem) and ABT-888 (1 μM; Enzo LifeSciences) for 3 h before LPS (100 ng/ml; Sigma-Aldrich, St. Louis, MO) stimulation. RAW 264.7 macrophages (Raw cells) stably downregulating ARTD1 were generated using viral transduction. Virus expressing an shARTD1 construct were generated in HEK293T cells. Then, 10 μg of shARTD1 plasmid, 6 μg of packaging plasmid, and 3.5 μg of viral envelope plasmid were transfected into HEK293T cells (4 × 106 cells per 10-cm plate) using calcium phosphate. The medium was changed at 6 to 8 h posttransfection, and supernatant containing virus was collected, centrifuged, and filtered (0.45-μm-pore-size cellulose acetate filters) after 3 days. RAW 264.7 cells were seeded 1 day prior to transduction on a 6-well plate (5 × 105 cells per well). Polybrene was added to a final concentration of 4 μg/ml overnight. Subsequently, 1 ml of supernatant containing virus was added per well. The medium was replaced at 8 h postinfection and changed to selective medium after 2 days (puromycin, 2 μg/ml; Invivogen, San Diego, CA). Cells were kept under constant selection.
siRNA transfection.
Negative-control AllStars (siMOCK), human siPARP1#6, mouse siPARP1#7, mouse siMLL1#1, mouse siSet7#1, and mouse siRelA#2 were ordered from Qiagen (Hilden, Germany). Cells were seeded at 50% confluence (2 × 105 cells per well) and transfected with 20 nmol of small interfering RNA (siRNA) per well (in six-well plate) with RNAiMAX Lipofectamine (Invitrogen). The experiment was performed 3 days after transfection.
RNA extraction and quantitative real-time PCR (qPCR) analysis.
RNA extraction was performed with the NucleoSpin RNA II kit (Macherey-Nagel, Düren, Germany). RNA was quantified with a NanoDrop (Thermo-Fisher Scientific, Waltham, MA), and 2 μg of RNA was reverse transcribed according to the supplier's protocol (high-capacity cDNA reverse transcription kit; Applied Biosystems, Foster City, CA).
qPCRs were performed witha SYBR green SensiMix SYBR Hi-ROX kit (Bioline Reagents, Ltd., London, United Kingdom) and a Rotor-Gene Q 2plex HRM system (Qiagen). See Table S1 in the supplemental material for primer sequences. The relative amounts of each mRNA were normalized to the RPS12 (mouse) and RPL28 (human) housekeeping genes.
Cell lysis, SDS-PAGE, and Western blot analysis.
Whole-cell extracts were prepared directly on plate by using a Tris lysis buffer (50 mM Tris [pH 8], 500 mM NaCl, 1% Triton X-100, 1 μg of pepstatin/ml, 1 μg of bestatin/ml, 1 μg of leupeptin/ml, 2 mM phenylmethylsulfonyl fluoride). Lysates were homogenized for 10 min at 4°C, followed by a 10-min centrifugation (maximum speed at 4°C) to eliminate cell debris. The protein concentration was quantified by a Bradford assay (Bio-Rad Laboratories, Hercules, CA), and 30 μg of protein extract was separated on a 10 or 7.5% SDS-polyacrylamide gel (120 V). The gel was blotted onto a polyvinylidene difluoride membrane and analyzed using protein-specific antibodies.
Coimmunoprecipitation.
Cells were harvested with a scraper and washed once with phosphate-buffered saline (PBS) for 5 min at 900 × g. The pellet was resuspended in 400 μl of hypotonic buffer (0.5% NP-40, 85 mM KCl, 5 mM HEPES [pH 7.4]) and directly centrifuged for 10 min at 6,200 × g using a cold centrifuge. Next, the pellet was resuspended in 200 μl of nuclear extraction buffer (50 mM Tris-HCl [pH 7.5], 150 mM KCl, 5 mM MgCl2, 0.2 mM EDTA, 20% glycerol, 0.1% NP-40) and sonicated twice for 30 s. The nuclear extract was incubated for 30 min at 4°C with 1 μl of DNase and sonicated again for 30 s, followed by a 10-min centrifugation (4°C and 3,500 × g). Proteins were quantified by using a Bradford assay. Immunoprecipitation was carried out with 300 μg of extract at 4°C overnight with 10 μl of monoclonal antihemagglutinin (anti-HA)-agarose beads (Sigma-Aldrich). After overnight incubation, the beads were washed three times with washing buffer (20 mM Tris-HCl [pH 7.5], 0.1 M KCl, 5 mM MgCl2, 0.2 mM EDTA, 10% glycerol, 0.1% Tween), resuspended in 2× Laemmli buffer (20 μl), and boiled for 5 min at 95°C. SDS-PAGE and Western blot analysis were performed as described above.
Immunofluorescence microscopy.
Cells were cultured on sterile coverslips (105 cells per well in a 24-well-plate) and grown overnight. After treatment with or without H2O2 (at 1 mM in fetal calf serum-free medium for 10 min), the cells were fixed (methanol-acetic acid [3:1], 5 min on ice) and washed twice with PBS. The cells were blocked for 30 min in PBS containing 5% milk powder and 0.05% Tween and incubated with 10H PAR antibody (1:350) in the same buffer (1 h at room temperature). Coverslips were incubated with secondary Cy3-conjugated antibody (for 1 h at room temperature in the dark). After being washed with PBS, the coverslips were mounted with Vectashield containing DAPI (4′,6′-diamidino-2-phenylindole; Vector Laboratories, Burlingame, CA). Conventional microscopy was carried out using a Leica DMI 6000B light microscope (Leica Microsystems GmbH, Wetzlar, Germany).
ChIP.
Chromatin immunoprecipitation (ChIP) analysis for H3, H3K4me3 and ARTD1 was performed as described previously (54) using magnetic Dynabeads (Life Technologies, Carlsbad, CA). ChIP analysis for p65 was performed as described previously (50), using protein A-agarose-salmon sperm DNA beads (Millipore, Billerica, MA).
Luciferase assay.
Cells were transfected with a specific siRNA as described above (2 × 104 cells per well in a 24-well plate). At 1 day after transfection, the medium was changed, and different luciferase constructs were transfected using a TransIT-3T3 transfection kit (Mirus, Madison, WI). A luciferase assay was performed after 48 h using a dual-luciferase reporter assay system (Promega, Madison, WI) according to the supplier's protocol.
Antibodies.
The following antibodies were used: PARP1/ARTD1 (H-250 [rabbit]), PARP-1 (C2-10 [mouse]), and p65 (C-20 [rabbit]) from Santa Cruz Biotechnology, Inc. (Dallas, TX); tubulin (mouse) from Sigma-Aldrich; H3K4me3 (rabbit) from Millipore; and histone H3 (rabbit) from Abcam PLS (Cambridge, United Kingdom). MLL1/HRX was obtained from Millipore. Secondary Cy3-conjugated AffiniPure goat anti-mouse antibody was obtained from Jackson ImmunoResearch Laboratories (Suffolk, United Kingdom), and 10H PAR (mouse) antibody was prepared in-house.
RESULTS
ARTD1 negatively regulates LPS-induced IL-6 expression in a RelA-dependent manner.
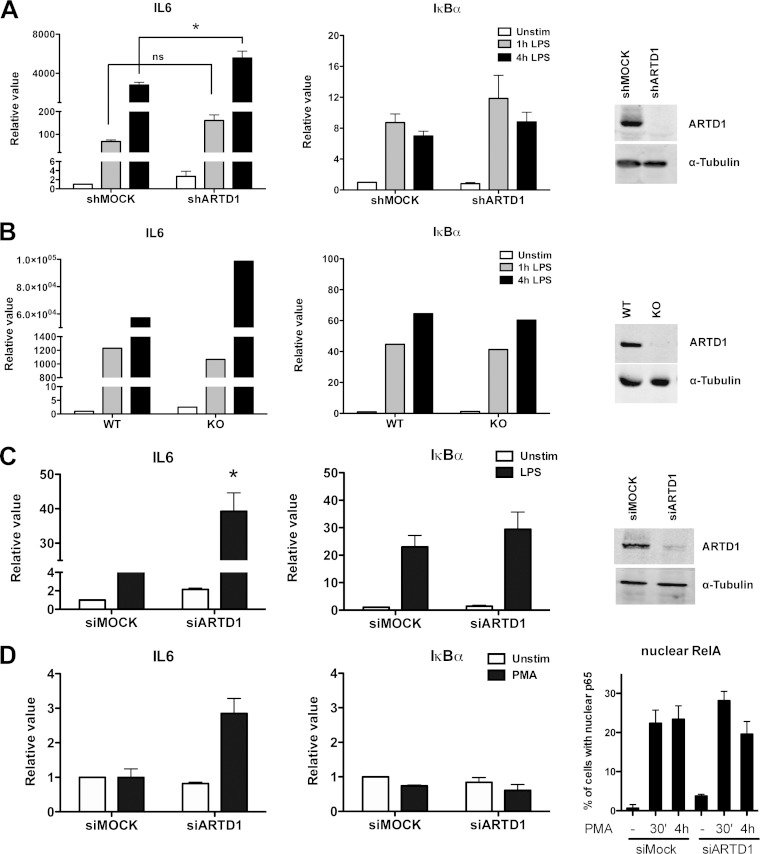
Stimulation of Raw cells with LPS for 1 or 4 h led to a strong induction of IL-6 transcript levels as identified by quantitative reverse transcription-PCR (RT-PCR) (Fig. 1A). In shARTD1-treated Raw cells (see Fig. 1A, right panel, for knockdown efficiency), IL-6 expression levels were comparably induced after 1 h but significantly enhanced at 4 h after LPS stimulation, suggesting that ARTD1 negatively regulates IL-6 gene expression at this later time point. The observed effect was specific for IL-6, since the NF-κB-dependent control gene IκBα was not affected by ARTD1 knockdown (Fig. 1A). Selectively enhanced IL-6 expression (compared to their respective controls) was also observed in LPS-stimulated primary BMDM from ARTD1 knockout mice (Fig. 1B) and NIH 3T3 fibroblasts treated with siARTD1 (Fig. 1C; see the right-hand panels for knockout and knockdown efficiencies), as well as in siARTD1-treated HEK293T cells stimulated with PMA (Fig. 1D; see the right-hand panel for the induction of RelA nuclear translocation), suggesting that the ARTD1-mediated negative regulation of IL-6 expression is cell type (and stimulus) independent.
FIG 1.
ARTD1 negatively regulates LPS- or PMA-induced IL-6 expression. IL-6 and IκBα gene expression was quantified by RT-PCR in Raw cells (A), BMDM (B), NIH 3T3 cells (C), and HEK293T cells (D). (A) IL-6 and IκBα expression upon LPS stimulation in Raw cells treated with shRNA specific against ARTD1 (n = 5). The right panel shows knockdown efficiency by Western blotting. (B) IL-6 and IκBα expression upon LPS stimulation in BMDM from wild-type and ARTD1 knockout mice (one representative experiment out of three). The right panel demonstrates knockout by Western blotting. (C) IL-6 and IκBα expression upon LPS stimulation in NIH 3T3 cells treated with siRNA specific against ARTD1 (n = 3 or 4). The right panel shows knockdown efficiency by Western blotting. (D) IL-6 and IκBα expression upon PMA stimulation in HEK293T cells treated with siRNA specific against ARTD1 (n = 2). The right panel demonstrates induction of p65 by PMA (quantitative representation of immunofluorescence analysis). The data are presented as means ± the standard deviations (SD) and were analyzed by one-way analysis of variance (ANOVA), followed by Bonferroni's post hoc test. *, P < 0.05.
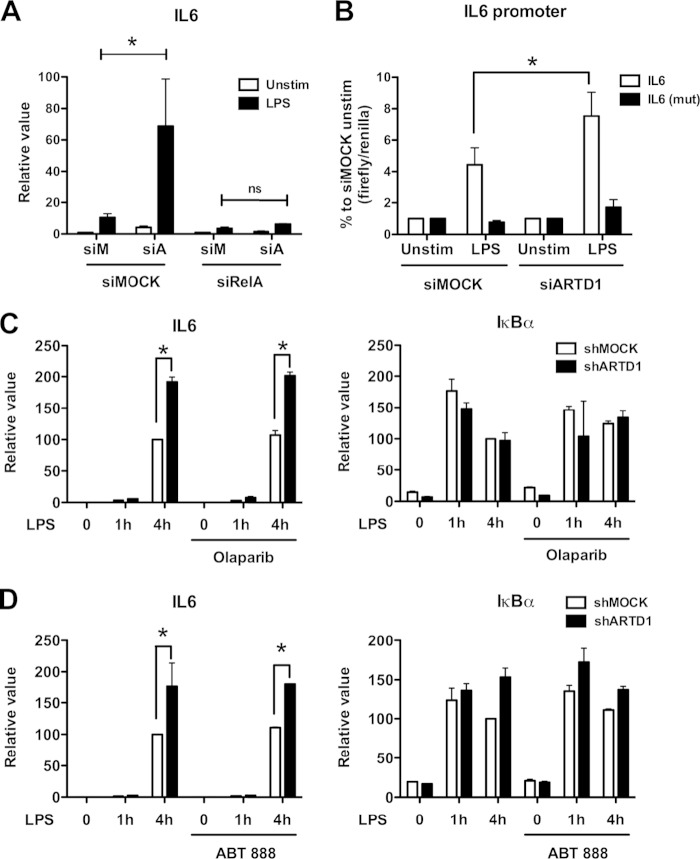
Because of their ease of transfectability, the mechanism of the negative regulatory effect of ARTD1 on IL-6 expression was further investigated using NIH 3T3 cells. To assess whether the negative regulation by ARTD1 was dependent on NF-κB, experiments with double knockdown of ARTD1 and RelA were performed. Enhanced IL-6 expression by ARTD1 knockdown in LPS-stimulated NIH 3T3 cells was completely abolished by concomitant downregulation of RelA (Fig. 2A), indicating that the negative regulatory effect of ARTD1 on IL-6 expression was highly dependent on the induction of RelA in these cells. To corroborate these results, reporter assays with a luciferase gene under the control of either the wild-type IL-6 promoter or a mutated IL-6 promoter lacking the NF-κB binding site were performed. In line with the analysis of the transcript levels, knockdown of ARTD1 led to enhanced IL-6 promoter transcriptional activity (Fig. 2B). This effect was not observed when the NF-κB binding site was mutated, confirming that the negative regulatory effect of ARTD1 under these conditions is also RelA-dependent and the chromatinization of the transfected reporter plasmids is sufficient to detect the ARTD1-mediated IL-6 repression.
FIG 2.
ARTD1 negatively regulates IL-6 expression in a p65-dependent manner and independent of its enzymatic activity. (A) IL-6 expression upon ARTD1 and RelA knockdown in NIH 3T3 cells stimulated with LPS or not stimulated (n = 3). (B) Expression of the luciferase reporter under the control of IL-6 promoter (after 4 h of LPS stimulation or unstimulated) in NIH 3T3 cells (n = 3). (C) IL-6 and IκBα expression upon olaparib treatment (1 μM) in Raw cells (n = 2). (D) IL-6 and IκBα expression upon ABT-888 treatment (1 μM) in Raw cells (n = 2). The data are presented as means ± the SD and were analyzed by one-way ANOVA, followed by Bonferroni's post hoc test. *, P < 0.05.
ARTD1 negatively regulates IL-6 expression independent of its enzymatic activity.
ARTD1 can regulate gene expression by ADP-ribosylating target proteins (including itself) or by its association with chromatin at the promoters of regulated genes (i.e., independently of its enzymatic activity). To determine whether the enzymatic activity of ARTD1 is required for the observed negative regulatory effect on IL-6 expression, Raw cells were stimulated with LPS in the absence or presence of the ADP-ribosylation inhibitor olaparib. Again, ARTD1 knockdown lead to enhanced IL-6 expression, whereas that of IκBα remained unaffected (Fig. 2C). Treatment of cells with olaparib, which effectively inhibited H2O2-induced PAR formation at the dose used for this experiment (data not shown), neither affected IL-6 expression in shMOCK cells nor enhanced expression in shARTD1 cells (Fig. 2C), indicating that ADP-ribosylation is not involved in the negative regulatory effect of ADRT1 on IL-6 expression. Comparable results were obtained with another ADP-ribosylation inhibitor, ABT-888 (Fig. 2D). Together, these results demonstrate that the observed derepression of IL-6 expression upon knockdown of ARTD1 is neither dependent on ARTD1-mediated ADP-ribosylation nor on that by another ARTD family member.
ARTD1 does not alter the H3 occupancy at the IL-6 promoter.
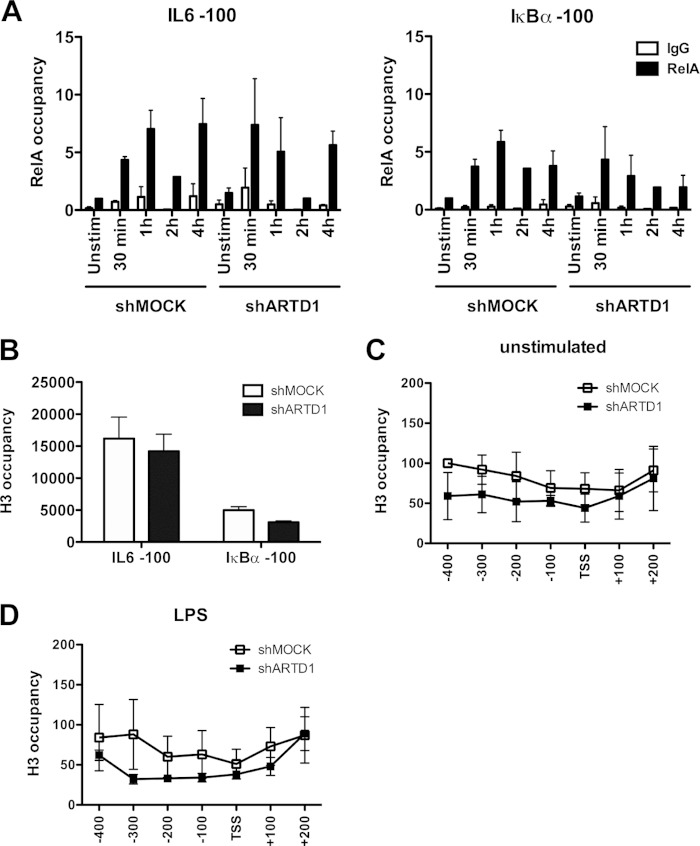
To investigate whether ARTD1 represses IL-6 expression indirectly by hampering RelA recruitment to the IL-6 promoter, LPS-induced recruitment of RelA to the IL-6 and IκBα promoters (100 bp upstream of the TSS was compared in shMOCK and shARTD1-treated Raw cells. The biphasic recruitment of RelA to the IL-6 promoter (i.e., immediate within 1 h and late at 4 h after stimulation) was not different between shMOCK and shARTD1-treated cells in regard to timing and extent (Fig. 3A). The ARTD1-dependent negative regulation of IL-6 expression was therefore not due to an effect on RelA recruitment to the IL-6 promoter.
FIG 3.
ARTD1 helps to maintain H3 at the IL-6 promoter. (A) Effect of ARTD1 knockdown on p65 occupancy on IL-6 and IκBα promoters upon LPS stimulation in Raw cells (n = 3). (B to D) H3 occupancy at the IL-6 and IκBα promoters in Raw cells under unstimulated conditions (shMOCK and shARTD1) (B and C) or after 4 h of LPS treatment (D) (n = 3). The data are presented as mean ± the SD.
Alternatively, as a chromatin-associated factor, ARTD1 may exert its regulatory function through the modulation of the chromatin state at the TSS of the IL-6 gene. To address the function of ARTD1 as a chromatin regulator at the promoter of IL-6, ChIP experiments were performed to analyze the H3 occupancy at the IL-6 and IκBα promoters. These analyses revealed that the chromatin at the IL-6 promoter was strongly enriched (at least 3-fold) in histone H3 compared to that at the IκBα promoter in shMOCK-treated cells (Fig. 3B), suggesting a more compact chromatin state at the IL-6 promoter (51). The H3 occupancy in shARTD1-treated Raw cells under basal conditions (i.e., untreated) was not significantly reduced compared to shMOCK-treated cells in the first 100 bp upstream of the TSS of the IL-6 promoter but also not further upstream and even downstream in the gene body (Fig. 3C). Upon LPS stimulation, the measured H3 content was also not significantly changed between shARTD1 and shMOCK cells (Fig. 3D), suggesting that ARTD1 affects IL-6 expression unlikely by altering the histone occupancy around the TSS.
ARTD1 regulates IL-6 expression through the H3K4me3 levels at the IL-6 promoter.
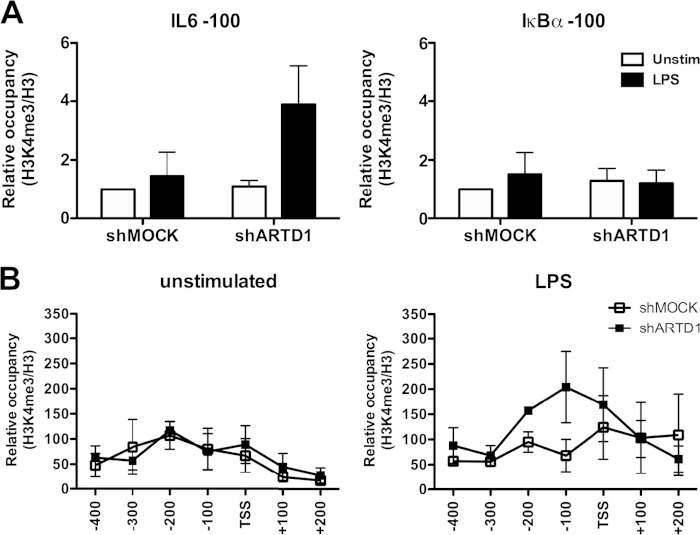
The results obtained thus far indicate that ARTD1 negatively regulates IL-6 expression independent of H3 recruitment or its enzymatic activity. However, ARTD1 may exert its regulatory function through the modulation of the histone modifications at the TSS of the IL-6 gene. The higher occupancy of H3 at the IL-6 promoter compared to the IκBα promoter (Fig. 3B) suggests that a H3 modification may be required to induce a more permissive chromatin state. H3K4me3 is a mark that is associated with active gene expression (52). We therefore elucidated whether ARTD1 inhibits H3K4me3 at the IL-6 promoter, thereby keeping the chromatin in a nonpermissive state. To study this possibility, ChIP experiments for H3K4me3 were performed and the IL-6 and IκBα promoters analyzed in detail. Upon ARTD1 knockdown and LPS stimulation, the relative occupancy of H3K4me3 100 bp upstream of the TSS of the IL-6 promoter was strongly increased, while no significant changes were observed at the IκBα promoter (Fig. 4A). More detailed analysis revealed that the strongest effects on relative H3K4me3 levels upon ARTD1 downregulation and LPS stimulation were observed between the TSS and 200 bp upstream (Fig. 4B), which corresponds to the location of the NF-κB response element. The increased H3K4me3 was observed at the 4-h time point, supporting the gene expression data and suggesting that ARTD1 represses H3K4me3 levels at the NF-κB binding site of the IL-6 promoter, thereby repressing IL-6 gene expression.
FIG 4.
ARTD1 knockdown is associated with increased H3K4me3 occupancy at the IL-6 promoter in LPS-stimulated cells. H3K4me3 occupancy at the IL-6 and IκBα promoters in Raw cells after 4 h of LPS stimulation (shMOCK and shARTD1) (n = 3) is shown.
MLL1 is responsible for the H3K4me3 at the IL-6 promoter and interacts with NF-κB.
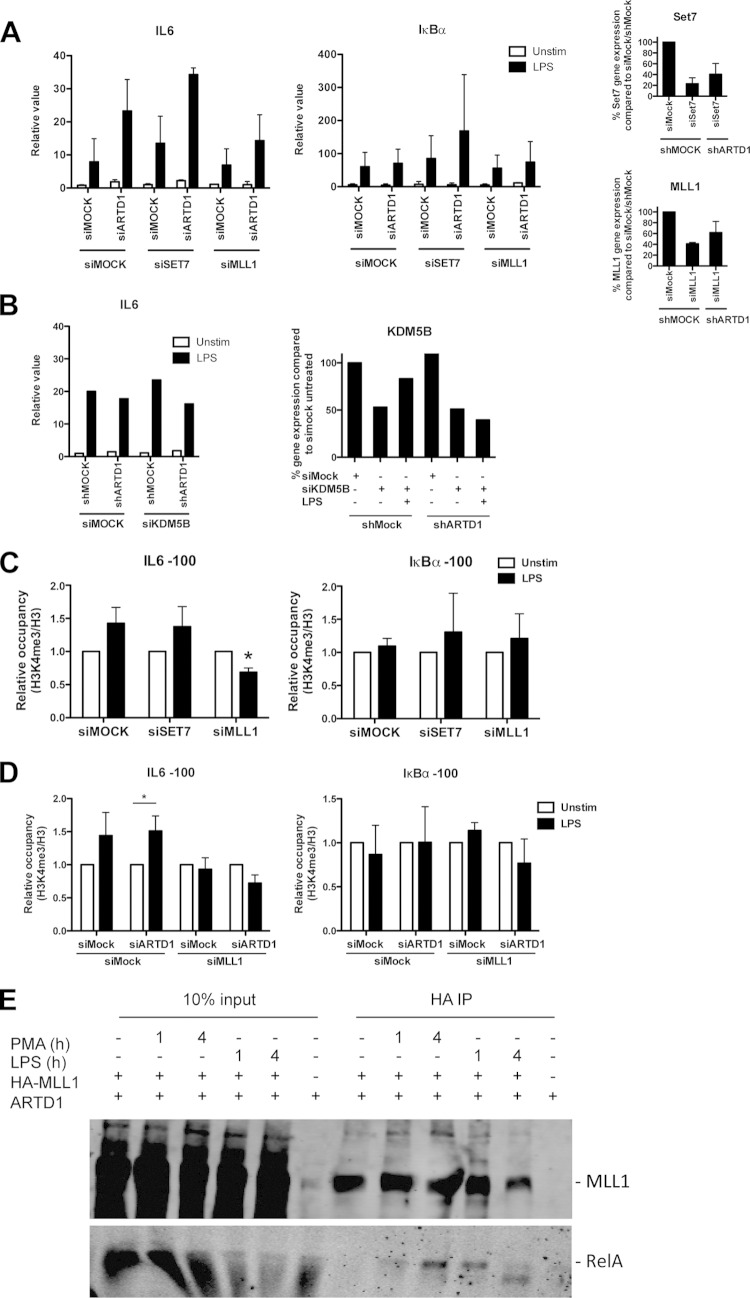
To identify the responsible H3K4 methyltransferase at the IL-6 TSS and to investigate a potential antagonistic effect with ARTD1, Set7/9 and MLL1 were knocked down by siRNA in NIH 3T3 cells in the presence or absence of ARTD1 (Fig. 5A; see the right-hand panels for the knockdown efficiencies). Whereas the enhanced IL-6 gene expression observed in siARTD1-treated cells was unaffected by knockdown of the methyltransferase Set7/9, it was rescued by knocking down MLL1, suggesting that MLL1 is the methyltransferase responsible for enhanced H3K4me3 formation at the IL-6 promoter upon knockdown of ARTD1 (Fig. 5A). The expression of IκBα was neither significantly affected by siARTD1 nor by siMLL1 treatment. The H3K4me3 could be also altered by a reduced activity of the histone demethylase KDM5B (43). However, knockdown of KDM5B did not lead to increased expression of IL-6 in either shMOCK or shARTD1 cells (Fig. 5B). Furthermore, knockdown of MLL1 but not of Set7/9 led to a significantly reduced H3K4me3 at the IL-6 promoter after LPS stimulation, whereas that at the control promoter IκBα was unaffected (Fig. 5C). These analyses provide strong evidence that the methyltransferase MLL1 and not Set7/9, or KDM5B regulates H3K4me3 at the IL-6 promoter in LPS-stimulated cells. The same observation was made when MLL1 was knocked down together with ARTD1, indicating that ARTD1 mediates its effect through MLL1 (Fig. 5D).
FIG 5.
MLL1 regulates IL-6 expression and H3K4me3 levels at the IL-6 promoter. (A) IL-6 and IκBα expression upon combined knockdown of ARTD1 and either SET7 or MLL1 in NIH 3T3 cells after 4 h of LPS stimulation (n = 3). The rightmost panels show the knockdown efficiency of siRNA treatment against SET7 and MLL1 by qPCR analysis. (B, left) IL-6 expression upon combined knockdown of ARTD1 and KDM5B in NIH 3T3 cells after 4 h of LPS stimulation (n = 1). (Right) Knockdown efficiency of siRNA treatment against KDM5B by qPCR analysis. (C) H3K4me3 occupancy at the IL-6 and IκBα promoters upon knockdown of ARTD1 and either SET7 or MLL1 in NIH 3T3 cells after 4 h of LPS stimulation (n = 3). The data are presented as means ± the SD and were analyzed by one-way ANOVA, followed by Bonferroni's post hoc test. *, P < 0.05. (D) H3K4me3 occupancy at the IL-6 and IκBα promoters upon combined knockdown of ARTD1 and either SET7 or MLL1 in NIH 3T3 cells after 4 h of LPS stimulation (n = 3). (E) MLL1 and p65 coimmunoprecipitation in HEK293T cells overexpressing MLL1 (lanes 1 and 4, unstimulated; lane 2, 1 h of PMA; lane 3, 4 h of PMA).
To test whether MLL1 forms a complex with NF-κB, complex formation was analyzed in HEK293T cells overexpressing HA-tagged MLL1. Since HEK293T cells lack TLR4 and therefore cannot be stimulated by LPS, cells were stimulated with phorbol myristate acetate (PMA). In HEK293T cells overexpressing MLL1, complex formation of MLL1 with endogenous RelA was detected in chromatin-free nuclear extracts 4 h after stimulation (Fig. 5E). Interestingly, the interaction between RelA and MLL1 was observed in a stimulus-dependent manner and at the same time point when the enhancement of IL-6 expression by ARTD1 knockdown was the most prominent. Due to the lack of suitable antibodies, the chromatin recruitment of MLL1 could not be analyzed, although others have recently reported that MLL1 is recruited to the chromatin in an NF-κB-dependent manner to regulate NF-κB-mediated gene expression (25).
ARTD1 forms a complex with MLL1 but leaves the IL-6 promoter in a stimulus-dependent manner.
To explain how ARTD1 interferes with MLL1-dependent IL-6 expression and H3K4me3 at the IL-6 promoter, MLL1 expression levels were analyzed. ARTD1 knockdown did neither significantly affect MLL1 expression nor MLL1 protein levels (Fig. 6A and B), suggesting that ARTD1 may rather be involved in the recruitment of MLL1 to the chromatin or may influence specific protein-protein interactions.
FIG 6.
MLL1 interacts with ARTD1 and p65 after LPS stimulation. (A and B) MLL1 gene (A) and protein (B) expression in shMOCK and shARTD1-treated Raw cells (n = 5 each). (C) MLL1 and ARTD1 coimmunoprecipitation in HEK293T cells overexpressing MLL1 and pretreated with siMOCK or siARTD1 (4 h PMA). (D) ChIP analysis of ARTD1 occupancy on IL-6 and IκBα promoters in Raw cells (n = 4).
To test, whether ARTD1 forms a complex with MLL1, MLL1 was immunoprecipitated from HEK293T cells overexpressing MLL1 and stimulated for 4 h with PMA. An interaction of MLL1 with endogenous ARTD1 was indeed detected in a stimulus-dependent manner at the same time point as for RelA (i.e., 4 h after stimulation) (Fig. 6C). The detected signal was specific, because ARTD1 knockdown obliterated the signal. Since the negative regulation of IL-6 expression by ARTD1 could not be explained by an altered recruitment of RelA (Fig. 3A) and the chromatin recruitment of MLL1 could not be analyzed, we investigated whether the chromatin recruitment of ARTD1 is changed during LPS stimulation. ChIP experiments with LPS-stimulated Raw cells revealed that ARTD1 was time-dependently released from the promoters of both IL-6 and IκBα upon LPS stimulation (Fig. 6D), suggesting that LPS stimulation changes the occupancy of ARTD1 at different chromatin loci.
In summary, we describe MLL1 as a new transcriptional coactivator of NF-κB and an additional molecular mechanism by which ARTD1 coregulates NF-κB-dependent transcription, namely, by the regulation of H3K4me3 through MLL1 and independent of ARTD1's enzymatic activity.
DISCUSSION
ADP-ribosylation and in particular ARTD1 have been implicated in many different and distinct cellular and biological processes (31). One of the most important functions of ARTD1 is the coregulation of inflammatory gene expression by direct modulation of transcriptional regulators or indirectly through alterations of the chromatin state (31).
Here, we elucidated the mechanism by which ARTD1 conveys a nonpermissive chromatin state at the IL-6 promoter, by interfering with MLL1-dependent H3K4me3 upon LPS stimulation. Interestingly, expression of the gene encoding the NF-κB inhibitor IκBα and other housekeeping genes was not ARTD1 dependent, which may be due to different chromatin architecture at the promoters of these two genes. The coregulatory function of ARTD1 was not mediated by its enzymatic activity.
The results presented here define the negative regulation of H3K4me3 at the IL-6 promoter by MLL1 as the mechanism by which ARTD1 modulates IL-6 expression. Previously, ARTD1 activity was shown to positively regulate transcription via the modification and inhibition of the histone demethylase KDM5B, responsible for the demethylation of H3K4me3 (43). In the present study, knockdown of MLL1 but not of SET7 or KDM5B altered the H3K4me3 of the IL-6 promoter. It is tempting to speculate that ARTD1, depending on the cellular context, its stimulation, and the availability of its substrate NAD+, can either repress H3K4me3, through ADP-ribosylation-independent binding to MLL1, or enhance H3K4me3 and therefore gene expression through ADP-ribosylation of KDM5B.
We observed that ARTD1 chromatin association is reduced upon LPS treatment after 4 h. Based on our recent studies (49), it is likely that the activation of the inflammasome at this time point is responsible for the proteolytic cleavage of ARTD1 at D214 and its subsequent release from chromatin. Since LPS itself is not expected to activate the inflammasome but to induce the expression of its components, it remains to be elucidated by which mechanism the inflammasome is activated.
According to our study, ARTD1 is bound to the IL-6 promoter even before stimulation of the cells, likely regulating the compaction and basal expression levels of IL-6. Similar to this result, biochemical studies with reconstituted chromatin have shown that in the absence of NAD+, ARTD1 promotes chromatin compaction independently of its enzymatic activity (53).
Our findings suggest a model in which ARTD1 represses IL-6 expression by interfering with the MLL1-induced H3K4me3 at the IL-6 promoter. Upon stimulation of cells, ARTD1 leaves the IL-6 promoter in a time-dependent manner. Interestingly, ARTD1 forms a complex with MLL1 only at the later time point (4 h, second NF-κB wave), when complex formation of MLL1 with NF-κB is also observed, suggesting that upon eviction of ARTD1, ARTD1 might compete with NF-κB for binding to MLL1, thus allowing only a certain amount of MLL1 to bind to NF-κB and thus damping the MLL1-induced H3K4me3 levels at this later time point. In contrast, in cells lacking ARTD1 or expressing reduced levels of ARTD1, all MLL1 binds to NF-κB and remains associated with the chromatin, resulting in increased H3K4me3 at the IL-6 promoter and consequently, increased IL-6 gene expression.
Since we were not able to investigate and quantify the recruitment of MLL1 to the IL-6 promoter, and the coimmunoprecipitation of MLL1 with NF-κB and/or ARTD1 does not allow us to distinguish whether all three proteins are found in the same complex or in distinct complexes containing MLL1 only associated with NF-κB or ARTD1, additional investigations are required to fully dissect the mechanism by which ARTD1 regulates MLL-1-driven NF-κB-dependent gene expression of IL-6, including also in developmental processes, homeobox gene expression or the development of acute leukemia. In this context, it will be of particular interest to investigate the regulation of IL-6 expression by ARTD1 in cells harboring MLL1 mutations.
In summary, our experiments have identified the dampening effect of ARTD1 on MLL1-dependent H3K4me3 as a new mechanism regulating the expression of the inflammatory cytokine IL-6. Taken together, these data strongly indicate that ARTD1 orchestrates chromatin accessibility and the posttranslational modification of histones indirectly through the interaction with the H3K4 methyltransferase MLL1 and thereby modulates the expression of IL-6. This apparently occurs in an ADP-ribosylation-independent manner. These results not only have important implications for our understanding and future analysis of MLL1 functions and IL-6 regulation but also highlight a potential cross talk between IL-6- and MLL1-induced pathologies.
Supplementary Material
ACKNOWLEDGMENTS
We thank Robert Roeder (The Rockefeller University, New York, NY) for providing the Flag-HA-tagged MLL1 pcDNA5 clone. We thank Florian Freimoser and Stephan Christen (Institute of Veterinary Biochemistry and Molecular Biology, University of Zurich, Zurich, Switzerland) for editorial assistance and critical input during the writing.
This study was supported in part by the Swiss National Science Foundation (grants 310030B_138667 and 310030_157019), the Kanton of Zurich, and the Novartis Foundation (to M.O.H.).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00196-15.
REFERENCES
- 1.Nishimoto N, Kishimoto T. 2006. Interleukin 6: from bench to bedside. Nat Clin Pract Rheumatol 2:619–626. doi: 10.1038/ncprheum0338. [DOI] [PubMed] [Google Scholar]
- 2.Ganapathi MK, Weizer AK, Borsellino S, Bukowski RM, Ganapathi R, Rice T, Casey G, Kawamura K. 1996. Resistance to interleukin 6 in human non-small cell lung carcinoma cell lines: role of receptor components. Cell Growth Differ 7:923–929. [PubMed] [Google Scholar]
- 3.Yamaji H, Iizasa T, Koh E, Suzuki M, Otsuji M, Chang H, Motohashi S, Yokoi S, Hiroshima K, Tagawa M, Nakayama T, Fujisawa T. 2004. Correlation between interleukin 6 production and tumor proliferation in non-small cell lung cancer. Cancer Immunol Immunother 53:786–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knupfer H, Preiss R. 2007. Significance of interleukin-6 (IL-6) in breast cancer. Breast Cancer Res Treat 102:129–135. doi: 10.1007/s10549-006-9328-3. [DOI] [PubMed] [Google Scholar]
- 5.Bromberg J, Wang T. 2009. Inflammation and cancer: IL-6 and STAT3 complete the link. Cancer Cell 15:79–80. doi: 10.1016/j.ccr.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iliopoulos D, Hirsch HA, Wang G, Struhl K. 2011. Inducible formation of breast cancer stem cells and their dynamic equilibrium with non-stem cancer cells via IL-6 secretion. Proc Natl Acad Sci U S A 108:1397–1402. doi: 10.1073/pnas.1018898108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hugo HJ, Lebret S, Tomaskovic-Crook E, Ahmed N, Blick T, Newgreen DF, Thompson EW, Ackland ML. 2012. Contribution of fibroblast and mast cell (afferent) and tumor (efferent) IL-6 effects within the tumor microenvironment. Cancer Microenviron 5:83–93. doi: 10.1007/s12307-012-0098-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raz Y, Erez N. 2013. An inflammatory vicious cycle: fibroblasts and immune cell recruitment in cancer. Exp Cell Res 319:1596–1603. doi: 10.1016/j.yexcr.2013.03.022. [DOI] [PubMed] [Google Scholar]
- 9.Servais C, Erez N. 2013. From sentinel cells to inflammatory culprits: cancer-associated fibroblasts in tumour-related inflammation. J Pathol 229:198–207. doi: 10.1002/path.4103. [DOI] [PubMed] [Google Scholar]
- 10.Armenante F, Merola M, Furia A, Palmieri M. 1999. Repression of the IL-6 gene is associated with hypermethylation. Biochem Biophys Res Commun 258:644–647. doi: 10.1006/bbrc.1999.0566. [DOI] [PubMed] [Google Scholar]
- 11.Armenante F, Merola M, Furia A, Tovey M, Palmieri M. 1999. Interleukin-6 repression is associated with a distinctive chromatin structure of the gene. Nucleic Acids Res 27:4483–4490. doi: 10.1093/nar/27.22.4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dandrea M, Donadelli M, Costanzo C, Scarpa A, Palmieri M. 2009. MeCP2/H3meK9 are involved in IL-6 gene silencing in pancreatic adenocarcinoma cell lines. Nucleic Acids Res 37:6681–6690. doi: 10.1093/nar/gkp723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takeuch O, Akira S. 2011. Epigenetic control of macrophage polarization. Eur J Immunol 41:2490–2493. doi: 10.1002/eji.201141792. [DOI] [PubMed] [Google Scholar]
- 14.Tang B, Zhao R, Sun Y, Zhu Y, Zhong J, Zhao G, Zhu N. 2011. Interleukin-6 expression was regulated by epigenetic mechanisms in response to influenza virus infection or dsRNA treatment. Mol Immunol 48:1001–1008. doi: 10.1016/j.molimm.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 15.Li Q, Verma IM. 2002. NF-κB regulation in the immune system. Nat Rev Immunol 2:725–734. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- 16.Karin M, Greten FR. 2005. NF-κB: linking inflammation and immunity to cancer development and progression. Nat Rev Immunol 5:749–759. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- 17.Tak PP, Firestein GS. 2001. NF-κB: a key role in inflammatory diseases. J Clin Invest 107:7–11. doi: 10.1172/JCI11830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hassa PO, Haenni SS, Buerki C, Meier NI, Lane WS, Owen H, Gersbach M, Imhof R, Hottiger MO. 2005. Acetylation of poly(ADP-ribose) polymerase-1 by p300/CREB-binding protein regulates coactivation of NF-κB-dependent transcription. J Biol Chem 280:40450–40464. doi: 10.1074/jbc.M507553200. [DOI] [PubMed] [Google Scholar]
- 19.Mao X, Gluck N, Li D, Maine GN, Li H, Zaidi IW, Repaka A, Mayo MW, Burstein E. 2009. GCN5 is a required cofactor for a ubiquitin ligase that targets NF-κB/RelA. Genes Dev 23:849–861. doi: 10.1101/gad.1748409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smale ST. 2010. Selective transcription in response to an inflammatory stimulus. Cell 140:833–844. doi: 10.1016/j.cell.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Wittwer T, Weber A, Schneider H, Moreno R, Maine GN, Kracht M, Schmitz ML, Burstein E. 2012. Regulation of NF-κB activity by competition between RelA acetylation and ubiquitination. Oncogene 31:611–623. doi: 10.1038/onc.2011.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sen N, Paul BD, Gadalla MM, Mustafa AK, Sen T, Xu R, Kim S, Snyder SH. 2012. Hydrogen sulfide-linked sulfhydration of NF-κB mediates its antiapoptotic actions. Mol Cell 45:13–24. doi: 10.1016/j.molcel.2011.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ansari KI, Mandal SS. 2010. Mixed lineage leukemia: roles in gene expression, hormone signaling and mRNA processing. FEBS J 277:1790–1804. doi: 10.1111/j.1742-4658.2010.07606.x. [DOI] [PubMed] [Google Scholar]
- 24.Hubert A, Henderson JM, Ross KG, Cowles MW, Torres J, Zayas RM. 2013. Epigenetic regulation of planarian stem cells by the SET1/MLL family of histone methyltransferases. Epigenetics 8:79–91. doi: 10.4161/epi.23211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, Zhu K, Li S, Liao Y, Du R, Zhang X, Shu HB, Guo AY, Li L, Wu M. 2012. MLL1, a H3K4 methyltransferase, regulates the TNFα-stimulated activation of genes downstream of NF-κB. J Cell Sci 125:4058–4066. doi: 10.1242/jcs.103531. [DOI] [PubMed] [Google Scholar]
- 26.Marschalek R. 2010. Mixed lineage leukemia: roles in human malignancies and potential therapy. FEBS J 277:1822–1831. doi: 10.1111/j.1742-4658.2010.07608.x. [DOI] [PubMed] [Google Scholar]
- 27.Zhang P, Bergamin E, Couture JF. 2013. The many facets of MLL1 regulation. Biopolymers 99:136–145. doi: 10.1002/bip.22126. [DOI] [PubMed] [Google Scholar]
- 28.Ramirez-Carrozzi V, Braas D, Bhatt D, Cheng C, Hong C, Doty K, Black J, Hoffmann A, Carey M, Smale S. 2009. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell 138:114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iliopoulos D, Hirsch H, Struhl K. 2009. An epigenetic switch involving NF-κB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell 139:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Korkaya H, Liu S, Wicha MS. 2011. Regulation of cancer stem cells by cytokine networks: attacking cancer's inflammatory roots. Clin Cancer Res 17:6125–6129. doi: 10.1158/1078-0432.CCR-10-2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kraus WL, Hottiger MO. 2013. PARP-1 and gene regulation: progress and puzzles. Mol Aspects Med 34:1109–1123. doi: 10.1016/j.mam.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 32.Rosenthal F, Hottiger MO. 2014. Identification of ADP-ribosylated peptides and ADP-ribose acceptor sites. Front Biosci (Landmark ed) 19:1041–1056. doi: 10.2741/4266. [DOI] [PubMed] [Google Scholar]
- 33.Elser M, Borsig L, Hassa PO, Erener S, Messner S, Valovka T, Keller S, Gassmann M, Hottiger MO. 2008. Poly(ADP-ribose) polymerase 1 promotes tumor cell survival by coactivating hypoxia-inducible factor-1-dependent gene expression. Mol Cancer Res 6:282–290. doi: 10.1158/1541-7786.MCR-07-0377. [DOI] [PubMed] [Google Scholar]
- 34.D'Amours D, Desnoyers S, D'Silva I, Poirier G. 1999. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J 342:249–268. doi: 10.1042/0264-6021:3420249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kraus W, Lis J. 2003. PARP goes transcription. Cell 113:677–683. doi: 10.1016/S0092-8674(03)00433-1. [DOI] [PubMed] [Google Scholar]
- 36.de Murcia G, Huletsky A, Lamarre D, Gaudreau A, Pouyet J, Daune M, Poirier G. 1986. Modulation of chromatin superstructure induced by poly(ADP-ribose) synthesis and degradation. J Biol Chem 261:7011–7017. [PubMed] [Google Scholar]
- 37.Kim M, Mauro S, Gévry N, Lis J, Kraus W. 2004. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell 119:803–814. doi: 10.1016/j.cell.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 38.Krishnakumar R, Gamble M, Frizzell K, Berrocal J, Kininis M, Kraus W. 2008. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science 319:819–821. doi: 10.1126/science.1149250. [DOI] [PubMed] [Google Scholar]
- 39.Altmeyer M, Messner S, Hassa PO, Fey M, Hottiger MO. 2009. Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites. Nucleic Acids Res 37:3723–3738. doi: 10.1093/nar/gkp229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Messner S, Altmeyer M, Zhao H, Pozivil A, Roschitzki B, Gehrig P, Rutishauser D, Huang D, Caflisch A, Hottiger MO. 2010. PARP1 ADP-ribosylates lysine residues of the core histone tails. Nucleic Acids Res 38:6350–6362. doi: 10.1093/nar/gkq463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ahel D, Horejsi Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I, Flynn H, Skehel M, West SC, Jackson SP, Owen-Hughes T, Boulton SJ. 2009. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science 325:1240–1243. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gottschalk A, Timinszky G, Kong S, Jin J, Cai Y, Swanson S, Washburn M, Florens L, Ladurner A, Conaway J, Conaway R. 2009. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc Natl Acad Sci U S A 106:13770–13774. doi: 10.1073/pnas.0906920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krishnakumar R, Kraus W. 2010. PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol Cell 39:736–749. doi: 10.1016/j.molcel.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hassa PO, Hottiger MO. 1999. A role of poly (ADP-ribose) polymerase in NF-κB transcriptional activation. Biol Chem 380:953–959. [DOI] [PubMed] [Google Scholar]
- 45.Hassa PO, Buerki C, Lombardi C, Imhof R, Hottiger MO. 2003. Transcriptional coactivation of nuclear factor-κB-dependent gene expression by p300 is regulated by poly(ADP)-ribose polymerase-1. J Biol Chem 278:45145–45153. doi: 10.1074/jbc.M307957200. [DOI] [PubMed] [Google Scholar]
- 46.Hassa PO, Covic M, Bedford MT, Hottiger MO. 2008. Protein arginine methyltransferase 1 coactivates NF-κB-dependent gene expression synergistically with CARM1 and PARP1. J Mol Biol 377:668–678. doi: 10.1016/j.jmb.2008.01.044. [DOI] [PubMed] [Google Scholar]
- 47.Hassa PO, Covic M, Hasan S, Imhof R, Hottiger MO. 2001. The enzymatic and DNA binding activity of PARP-1 are not required for NF-κB coactivator function. J Biol Chem 276:45588–45597. doi: 10.1074/jbc.M106528200. [DOI] [PubMed] [Google Scholar]
- 48.von Lukowicz T, Hassa PO, Lohmann C, Borén J, Braunersreuther V, Mach F, Odermatt B, Gersbach M, Camici GG, Stähli BE, Tanner FC, Hottiger MO, Lüscher TF, Matter CM. 2008. PARP1 is required for adhesion molecule expression in atherogenesis. Cardiovasc Res 78:158–166. doi: 10.1093/cvr/cvm110. [DOI] [PubMed] [Google Scholar]
- 49.Erener S, Petrilli V, Kassner I, Minotti R, Castillo R, Santoro R, Hassa PO, Tschopp J, Hottiger MO. 2012. Inflammasome-activated caspase 7 cleaves PARP1 to enhance the expression of a subset of NF-κB target genes. Mol Cell 46:200–211. doi: 10.1016/j.molcel.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 50.Saccani S, Pantano S, Natoli G. 2001. Two waves of nuclear factor κB recruitment to target promoters. J Exp Med 193:1351–1359. doi: 10.1084/jem.193.12.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 52.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. 2002. Active genes are tri-methylated at K4 of histone H3. Nature 419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 53.Wacker D, Ruhl D, Balagamwala E, Hope K, Zhang T, Kraus W. 2007. The DNA binding and catalytic domains of poly(ADP-ribose) polymerase 1 cooperate in the regulation of chromatin structure and transcription. Mol Cell Biol 27:7475–7485. doi: 10.1128/MCB.01314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Santoro R, Li J, Grummt I. 2002. The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat Genet 32:393–396. doi: 10.1038/ng1010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.