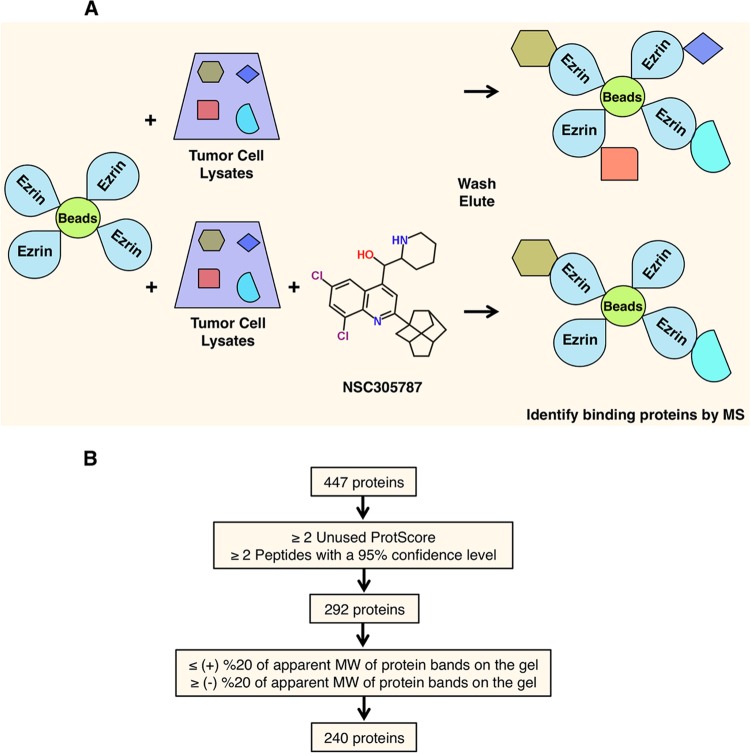
FIG 1.
Identification of candidate NSC305787-competed ezrin-binding proteins. (A) Schematic diagram describing an affinity pulldown coupled with an MS-MS approach for identification of ezrin-interacting proteins that can be competed away by NSC305787. Recombinant ezrin was purified and coupled with CNBr-activated Sepharose 4B beads. The ezrin-coated beads were then used as bait on total cell lysates from K7M2 mouse OS cells in the presence of either NSC305787 or vehicle control. The proteins bound to the beads were eluted by boiling samples in SDS-PAGE sample buffer. Proteins were then run on an SDS-PAGE gel and stained with Coomassie brilliant blue. Protein bands that were competed away with NSC305787 were analyzed by nano-LC–MS-MS. (B) Schematic flow diagram for analysis of the raw data obtained from MS. Through stepwise filtering, a total of 240 potential ezrin-interacting proteins that can be competed away by NSC305787 were identified. Proteins identified by MS-MS were first filtered based on the elimination of proteins that were not identified by at least two unique peptides with a confidence level of ≥95% and with a ProteinPilot Unused ProtScore of ≥2 (99% confidence level). Further subtraction of proteins was then based on the removal of proteins whose predicted molecular weights differed by more than 20% from the molecular weights of the corresponding bands on the gel.