Fig. 2.
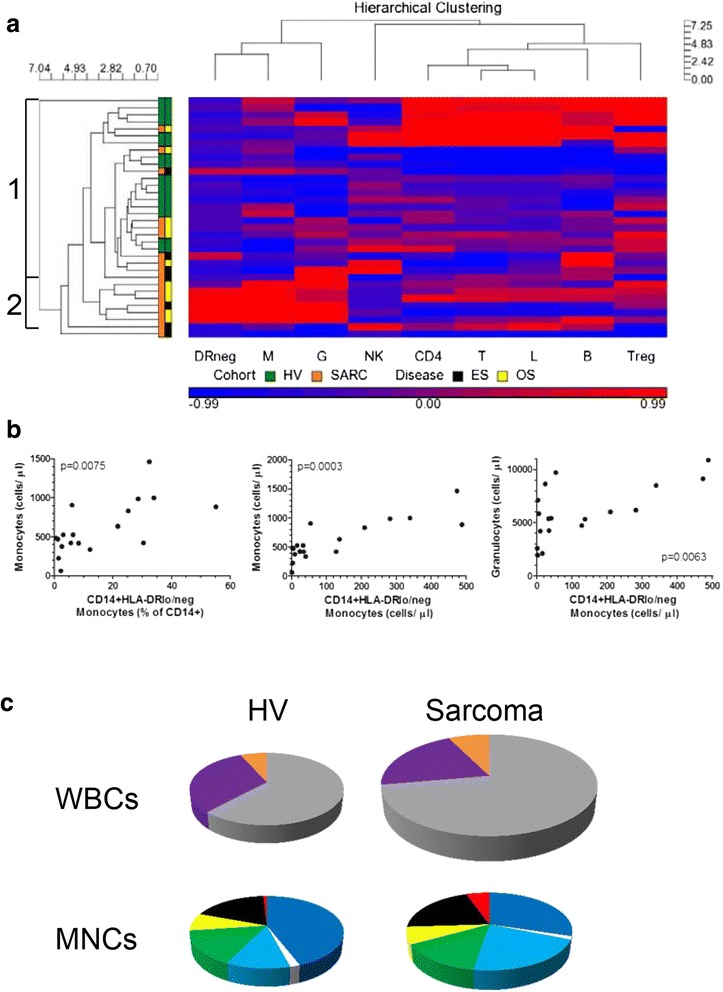
Hierarchical clustering of immune phenotypes reveals potential differential subgrouping of sarcoma patients based on compositional changes in peripheral blood immune phenotypes. a. Hierarchical clustering dendrogram showing the relationships of healthy volunteers and sarcoma patients based on nine immune phenotypes (DRneg- CD14+HLA-DRlo/neg monocytes; M- monocytes; G- granulocytes; NK- natural killer cells; CD4- CD4 T cells; T- T cells; B- B cells; Treg- CD4+CD25+CD127lo T cells). Profiles were defined by including a minimum of 5 individuals. Values shaded in red indicate values above the mean of the HV value and blue indicates values below. Two profiles of groups were identified. b. Hierarchical clustering also can reveal relationships of immune phenotypes to each other. The percentage and cell counts of CD14+HLA-DRlo/neg monocytes correlated to the cell counts of monocytes and the cell counts of CD14+HLA-DRlo/neg monocytes correlated to the cell counts of granulocytes. c. Illustrative comparisons of the compositional differences in healthy volunteers and sarcoma patients. For total white blood cells (WBCs); the percentages of granulocytes (gray), lymphocytes (purple), and monocytes (orange) are shown. The pie graphs are also sized based on cell counts where the sarcoma pie graph is sized in proportion to the HV pie graph. Mononuclear cells (MNCs) show CD4 T cells (dark blue), Tregs (white), CD8 T cells (light blue), B cells (green), NK cells (yellow), CD14+HLA-DR+ monocytes (black), and CD14+HLA-DRlo/neg monocytes (red). The sarcoma pie graph is sized in proportion to the HV graph
