Figure 4.
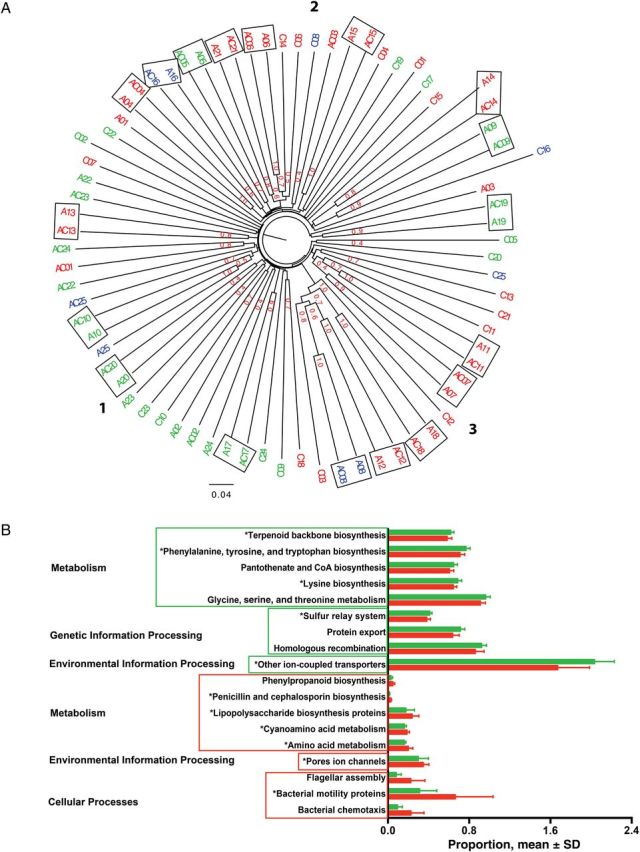
Clustered displays of taxonomic and functional data from 50 subjects. A, Unweighted pair group method with arithmetic mean cluster of the 75 samples from the 50 subjects, based on the unweighted Unifrac distance matrix with Jackknife analysis. The samples from the same microenvironment type are indicated by the same color, as follows: dry sites, red (n = 36); moist sites, green (n = 30); and sebaceous sites, blue (n = 9). The boxed samples are from the same subject. The 3 clusters are shown with bold numbers. In cluster 1, 20 of 27 samples (74%) are from moist skin. In cluster 3, 13 of 18 samples (72%) are from dry skin. Cluster 2 is a mixture of the 3 skin types. Jackknife support fractions of ≥0.5 are shown. B, Phylogenetic Investigation of Communities by Reconstruction of Unobserved States–predicted metagenome with significant differences in relative abundance. Samples are from patients with abscesses who did (n = 8; red) or did not (n = 17; green) receive antibiotics, and functional gene pathways categories with linear discriminant analysis scores of ≥2 are shown. Five of 9 functional gene pathways (56%) in both groups are constituents of the metabolism category. The red box indicates gene pathways with significant abundance in the patients with a history of relatively recent antibiotic use; the green box indicates significant abundance in the patients without recent antibiotic use history. *P < .05 for gene pathway categories that are significantly different after false-discovery rate correction. Abbreviation: SD, standard deviation.
