Figure 3.
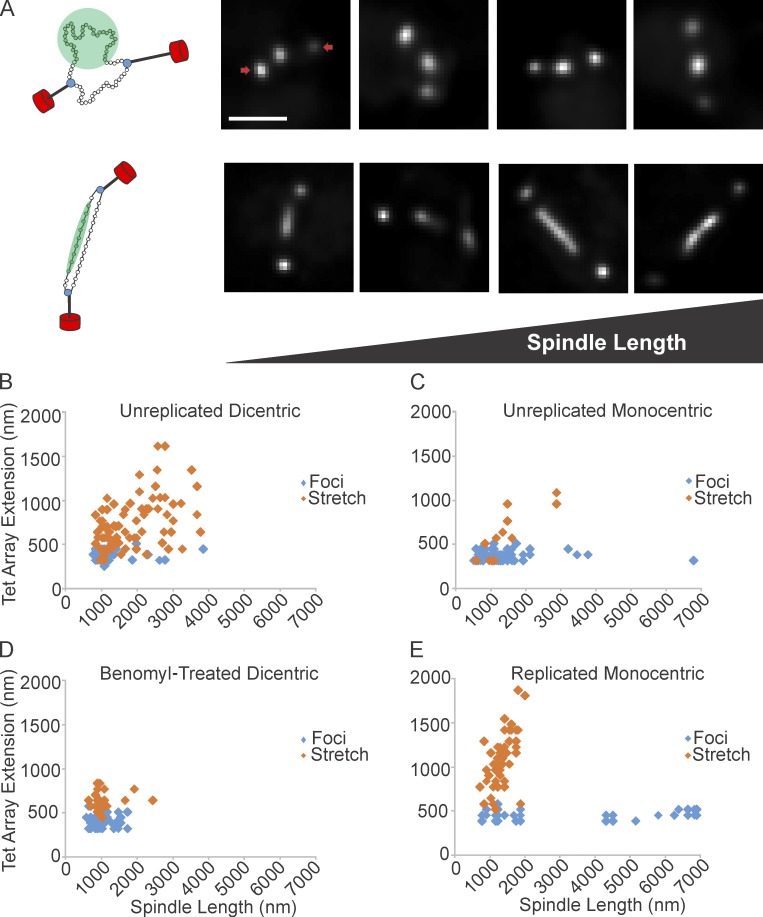
Tension along the spindle axis is sufficient to release nucleosomes. (A, left) Schematic of dicentric plasmid. The plasmid is attached to kMTs (black) via the kinetochore (blue). Nucleosomes are green, SPBs are red. The DNA appears as a near-diffraction spot (green circle, top) or a filament (green bar, below). Right four panels show deconvolved images of near-diffraction foci (top) and filaments (bottom) of dicentric plasmids. SPBs are indicated by arrows. (B) TetO/TetR-GFP extension vs. spindle length for unreplicated dicentric (n = 129 arrays). (C) Unreplicated monocentric plasmids (n = 102 arrays). (D) Benomyl-treated dicentric plasmids (n = 92 arrays). (E) Replicated monocentric plasmids (n = 94 arrays). Scatter plots display population data from multiple experiments. Bar, 1 µm.