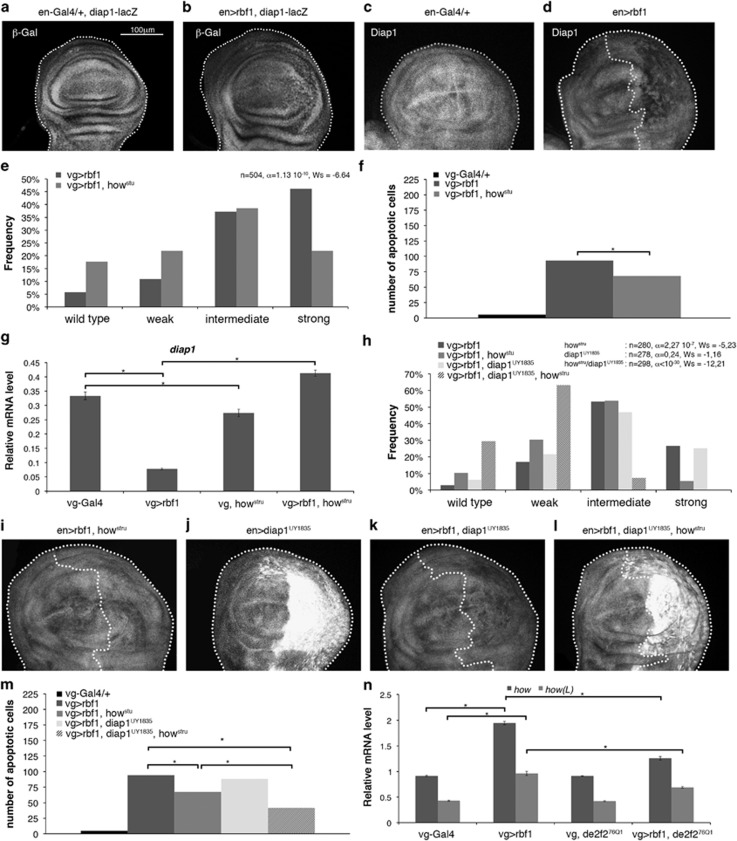
Figure 3.
Rbf1 and dE2F2 increases how mRNA leading to diap1 mRNA destabilization. (a) diap1-lacZ transgene was used to report diap1 transcription in en-gal4/+ and en-gal4>UAS-rbf1 genetic contexts. β-Gal immunostaining (white) of the discs are shown in (a, b). Diap1 immunostaining (white) are shown in (c, d) and (i–l). The genotypes are indicated at the top of the image. Posterior compartment of the wing disc (on the right) is delimitated by dotted line when rbf1 is overexpressed. All the pictures presented in Figure 3 are at the same scale, scale bar: 100 μm. Distribution of notch wing phenotypes in vg-Gal4>UAS-rbf1 and vg-Gal4>UAS-rbf1; howstru flies (e), vg-Gal4>UAS-rbf1, vg-Gal4>UAS-rbf1; howstru, vg-Gal4>UAS-rbf1; UAS-diap1UY1835 and vg-Gal4>UAS-rbf1; UAS-diap1UY1835; howstru flies (h). Wing phenotypes were grouped into four categories according to the number of notches (wild type, weak, intermediate, strong). Statistical analysis was performed using Wilcoxon tests. Each experiment was independently performed three times; as the results were similar, only one experiment is presented here. Quantification of TUNEL-positive cells (f and m) in the wing pouch of genotypes studied in (e) and (h). Asterisks indicate a statistically significant difference between two genotypes (Student's t-test, P<0.05). Quantification of diap1 (g), how and how(L) (n) mRNAs in wing imaginal discs by RT-qPCR. Data are normalized against rp49 and correspond to the mean of three independent experiments. Error bars are the S.E.M. Asterisks indicate statistically significant difference between two genotypes (Student's t-test, P<0.05)