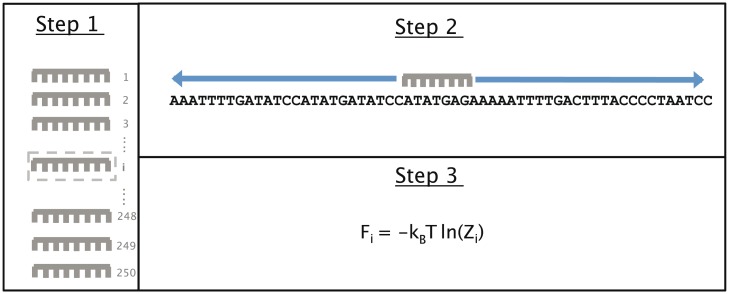
Fig 1. Cartoon illustrating our model for computing the free energy of nonconsensus protein-DNA binding.
Schematic representation of the procedure for computing the nonconsensus free energy. Step 1: In order to model nonspecific TF-DNA binding, we generate an ensemble of 250 random TFs. Step 2: Each TF moves within a sliding window of width L bp. The TF-DNA binding energy is computed at each location of TF along the sliding window using the random potential. Step 3: For each TF we calculate the TF-DNA binding free energy. We repeat this process for all random TFs and compute the average nonconsensus binding free energy with respect to this ensemble of random TFs. Moving the sliding window along the genome, we assign the nonconsensus TF-DNA binding free energy at each genomic location. We assume that each random binder makes contacts with M bps upon DNA binding. For each model TF (random binder), we define the partition function of protein-DNA binding within the chosen sliding window of width L bp. We used L = 50 bp (i.e. the sliding window size) in our calculations of the genome-wide nonconsensus TF-DNA binding free energy profiles, and L = 36 bp for calculations of the nonconsensus TF-DNA free energies for in vitro protein binding microarray (PBM) experiments. In the latter case, we do not move the sliding window since each DNA sequence in the PBM library is 36-bp long.