Figure 4.
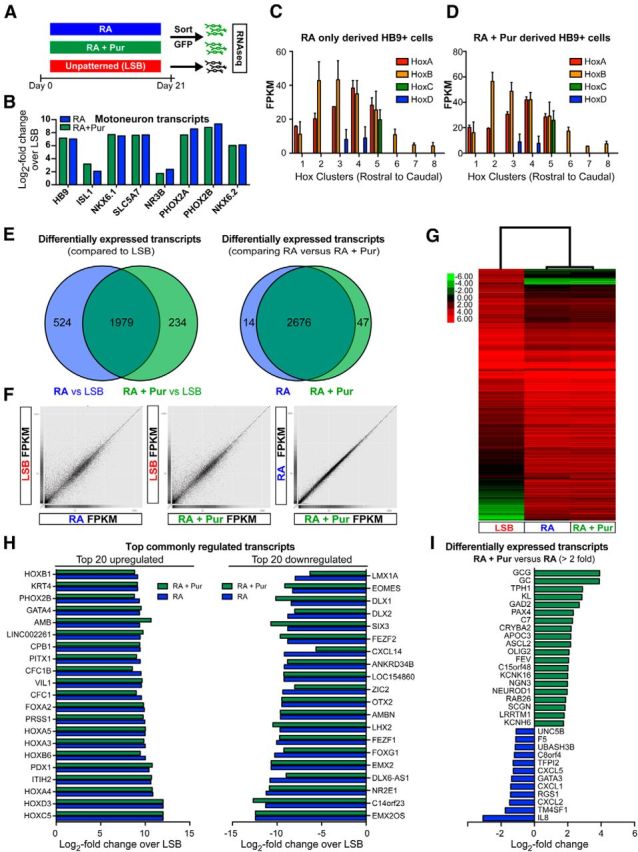
Transcriptome analysis of HB9::GFP-purified MNs derived by RA alone versus RA + Pur treatment. A, Schematic illustration depicting experimental paradigm for RNA sequencing. B, Selective expression of MN transcripts in isolated GFP+ cells derived with RA or RA + Pur compared with LSB. Hox gene expression demonstrates nearly identical anteroposterior identity of MNs derived in the absence (C) and presence (D) of extrinsic SHH. E, Comparison of genes differentially expressed between LSB and RA-alone- or RA + Pur-derived GFP+ populations shows significant overlap of MN populations. Of those differentially expressed genes compared with LSB, only 14 and 47 exhibit significant expression between RA-alone- and RA + Pur-derived MNs, demonstrating striking similarity between the populations. Scatterplots comparing FPKMs of individual genes (F) and hierarchical clustering of differentially expressed genes (G) further corroborate remarkable similarity between RA-alone- and RA + Pur-patterned MN populations. H, The top 20 commonly upregulated (left) and downregulated (right) genes expressed by RA- and RA + Pur-derived MNs compared with LSB. Genes are ranked by RA. I, Genes exhibiting significantly different (>2-fold) expression between RA + Pur and RA MN populations.
