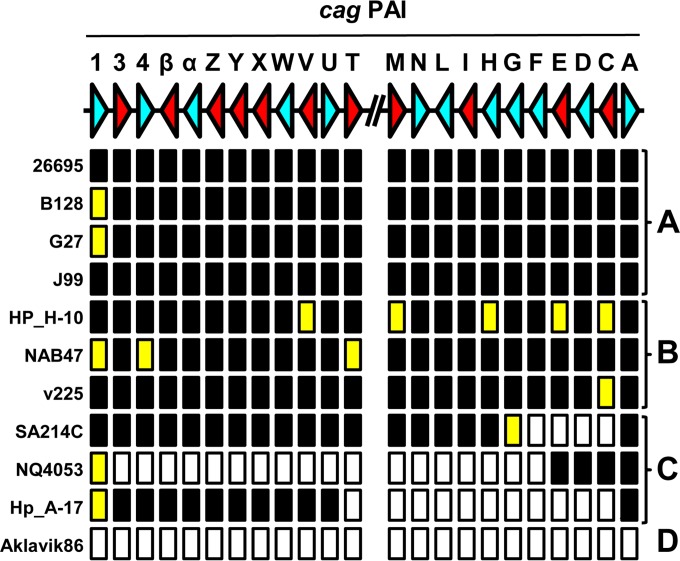
FIG 5.
Subdivision of presence or absence and functionality analysis of the H. pylori cag PAI. BLAST+ analysis of H. pylori genomes and encoded proteomes was used to determine whether all of the genes of the cag PAI were present or absent and, if present, whether they are predicted to encode a full-length protein. The top line shows a schematic representation of a gene map of the two arms of the cag PAI of H. pylori 26695, with each gene being represented by an arrowhead and letters representing the gene names. The direction of the arrowhead shows the transcriptional orientation. Genes essential for interleukin induction or pilus formation (38, 39) are red, whereas nonessential genes are light blue. The absence or presence of the gene and encoded protein are indicated below the gene map, with black rectangles indicating that both the gene and protein are present, yellow rectangles indicating the presence of the gene but absence or truncation of the corresponding protein, and white rectangles indicating the absence of the gene and protein. The H. pylori genomes could be subdivided in four classes based on the cag PAI status as follows: A, complete and functional cag PAI; B, complete cag PAI but functionally negative because of lack of expression of essential components; C, contains only fragments of the cag PAI, with large fragments missing; D, cag-negative genome lacking the cag PAI completely. Representative examples of each subgroup are shown. For full analyses of the individual genes of the cag PAI, see Table S4 in the supplemental material.