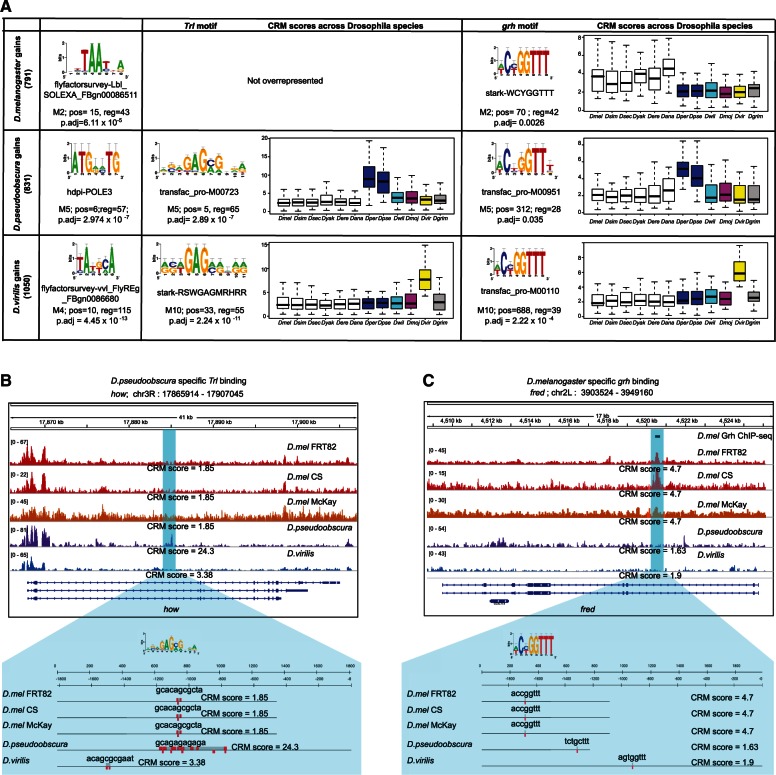
Fig. 6.
Lineage-specific Trl and Grh motifs are associated with lineage-specific open chromatin regions. (A) The rows represent FAIRE-Seq peak gains for each species. The first column shows the first and most significant PWM, for which CRM score changes overlap with open chromatin changes. The second column reports the position and P value of the Trl motif, and the third column the Grh motif. Trl and Grh CRM gains and losses correlate significantly with open chromatin changes. (B) Example of a Dpse-specific FAIRE-Seq peak, predicted to be gained because of a gain of a Trl CRM (increased CRM score from 1.85 to 24.3). (C) Example of a gene, fred, with a gain of a FAIRE-Seq peak in Drosophila melanogaster, predicted to be caused by a gain in Grh-binding sites. This position also contains a Grh ChIP-Seq peak (Potier et al. 2014).