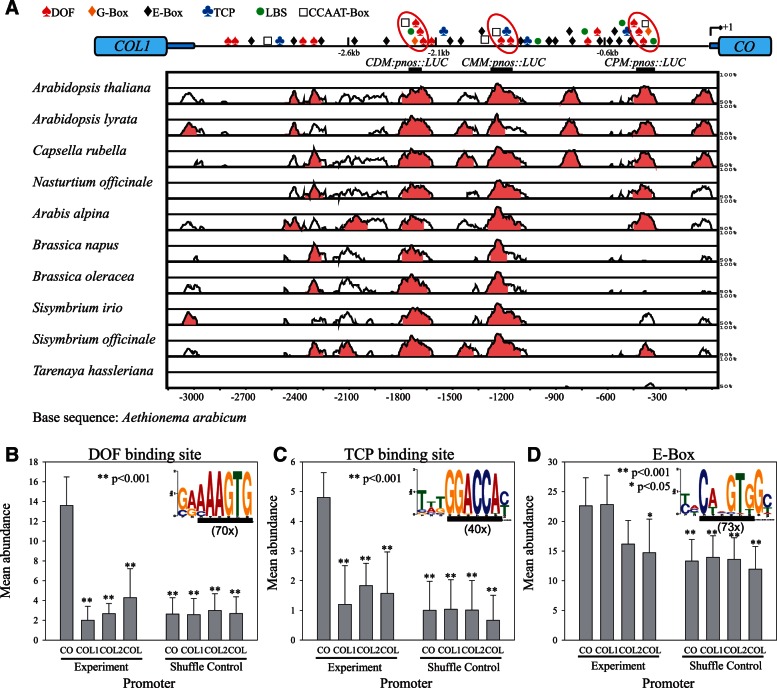
Fig. 4.
Conservation and cis-element composition of CO promoter in the Brassicaceae. (A) Pairwise alignment of the Aethionema arabicum CO promoter to orthologous sequences from ten Brassicaceae CO promoters as well as Tarenaya hassleriana COL promoter displayed as VISTA plots. Sequence similarity (%) was calculated in 100-bp sliding window while light-red color indicates greater than 75% identity. For clarity a diagram of Arabidopsis thaliana CO promoter is displayed above, indicating position of cis-elements (DOF = AAAGTG, TCP = GGACCA, E-Box = CANNTG, G-Box = CACGTG, LBS = GATWCG, and CCAAT-Box = CCAAT). Approximate positions of CO promoter sequences used for LUC reporter (CDM:pnos:LUC, CMM:pnos::LUC, CPM:pnos::LUC) and GFP:CO (2.6, 2.1, and 0.6 kb) complementation constructs in this study are indicated in relation to the vista plot. (B–D) Short motif search (MEME, 6–10 bp) in CO promoters from ten Brassicaceae species (see A) resulted in three elements that were found 70 (B), 40 (C), and 73 times (D), respectively (displayed as WEBLOGO). Identified motifs (underlined) contain the extended DOF binding site (B, AAAGTG), the TCP binding site (C, GGACCA), and the E-Box (D, CANNTG). Mean Abundance of these three cis-elements was determined in promoters (−4 kb) of Brassicaceae CO, COL1, and COL2 as well as non-Brassicaceae COL orthologs (COL) (B–D, Experiment, ±SEM). The same analysis was performed 100 times with shuffled data sets as control for base composition (Shuffle Control). Brassicaceae sequences include homologs from A. thaliana, A. alpina, Eutrema parvulum, Brassica rapa (1 × CO, 2 × COL1, and 2 × COL2), and Ae. arabicum (CO and COL2). Promoters from non-Brassicaceae species include T. hassleriana (ThCOL), Medicago truncatula (MtCOLa), Populus Trichocarpa (PtCO1; PtCO2), Solanum tuberosum (StCO1; StCO2), and Vitis vinifera (VvCO). Statistical analysis indicates significant differences (*P < 0.05; **P < 0.001) in mean abundance compared with CO.