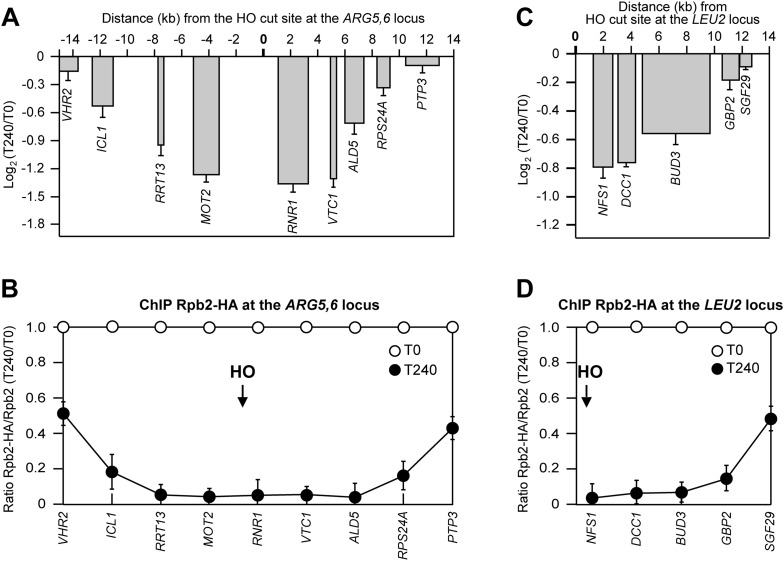
Figure 3. DSB-induced transcriptional inhibition at different chromosomal loci.
(A) YEPR exponentially growing cell cultures of tGI354 strain, carrying the HO site at the ARG5,6 locus and the deletion of RAD51 gene, were transferred to YEPRG at T0 to induce HO. RNA levels of genes located at different distances from the HO cut site were evaluated by qRT-PCR at T0 and T240 after HO induction, as described in Figure 2A. Results are presented as ratios between T240 and T0. The mean values ±s.d. are represented (n = 3). (B) Binding of Rpb2-HA from samples collected in (A) was evaluated by ChIP and qPCR as described in Figure 2B. Primers used were the same as in (A). The mean values ±s.d. are represented (n = 3). (C) YEPR exponentially growing cell cultures of YFP17 strain, carrying the HO cut site at the LEU2 locus, were transferred to YEPRG at T0 to induce HO. RNA levels were analyzed as in Figure 2A but normalized to the ALG9 gene transcript. Only transcription of genes located on the right side of the HO-induced DSB was analyzed because no transcription units are present on the left side of the break. The mean values ±s.d. are represented (n = 3). (D) Binding of Rpb2-HA from samples collected in (C) was evaluated by ChIP and qPCR as described in Figure 2B. Primers used were the same as in (C).