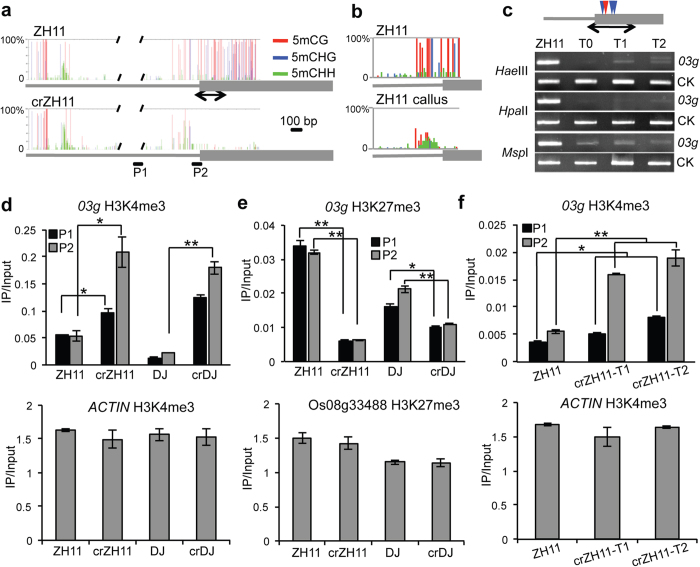
Figure 2. Stable 03g activation was associated with DNA methylation loss, histone H3K27me3 removal and H3K4me3 deposition.
(a) 03g was heavily methylated in CG and CHG contexts at transcription start site (TSS) and 5′ UTR in leaf tissues of normal wild type ZH11, but demethylated in leaf tissues of callus regenerated ZH11 (crZH11) plants. DNA methylation was determined by bisulfite sequencing. More than 20 clones for each genotype were sequenced. Solid gray line: upstream promoter region; grey bar: transcribed region of 03g gene. Primers sets (P1, P2) used in ChIP assays are indicated. Black slashes indicate the gap, which is not sequenced. (b) DNA methylation of 03g was lost in callus. Bisulfite sequencing region is represented by the double arrows shown in a. (c) DNA demethylation was maintained in subsequent generations, shown by enzyme digestion followed by PCR amplification. PCR amplification region was the same as bisulfite sequencing region in a. CK, control DNA fragments without enzyme recognition sites. Blue triangle indicates HpaII/MspI cut sites, red triangle indicates HaeIII cut sites. PCR products of different samples for each primer set were run in the same gel and cropped in the same picture. (d,e) H3K4me3 (d) and H3K27me3 (e) at promoter (P1) and TSS region (P2) of 03g were respectively increased and decreased in callus-regenerated (cr) plants of two different rice cultivars (ZH11 and DJ). (f) Increased H3K4me3 in 03g was maintained in subsequent T1 and T2 generations. ACTIN was used as a control for H3K4me3. Os08g33488, which is methylated by H3K27me315, was used as a control for H3K27me3. Student t-tests were performed from 3 biological repeats. *p < 0.05, **p < 0.01.