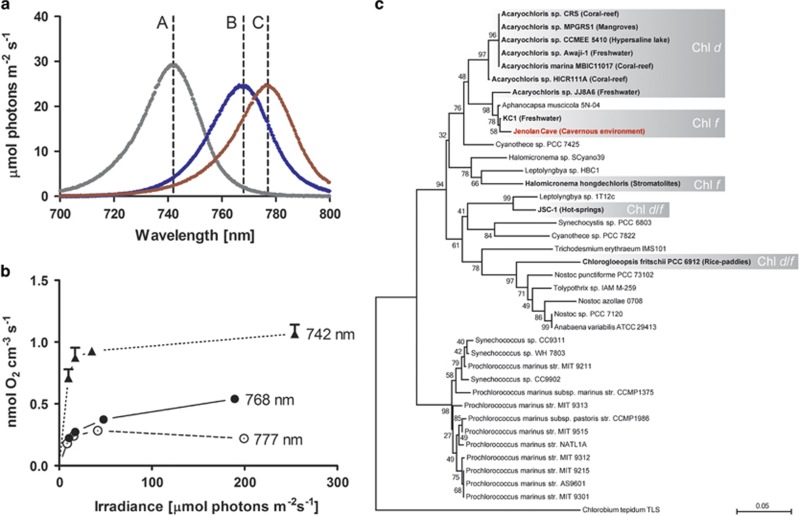
Figure 2.
Taxonomic affiliation and O2 evolution of Chl f-containing cells as determined by O2 microelectrode measurements and 16 S rDNA amplicon sequencing. (a) Emission spectra of narrow-band light-emitting diodes (LEDs) used in this study, with peak emissions at 742, 768 and 777 nm indicated by a–c, respectively. (b) Gross photosynthesis measured via an O2 microsensor placed in a clump of agarose-embedded Chl f-containing cells. Different NIR irradiance was administered by the LEDs in a and by altering the distance of the LEDs to the embedded cells. (c) Phylogenetic affiliation of known Chl f and/or Chl d-containing cyanobacteria (highlighted in gray) and their respective habitat/place of isolation. Taxonomy was determined by clustering all known oxygenic phototrophs found in enrichment cultures from this study (at order level) into a single OTU (=292 bp length, see Supplementary Materials for details). Phylogeny was inferred using Maximum-likelihood in conjunction with the GTR +I +G nucleotide substitution model, tree stability was tested using bootstrapping with 100 replicates. The analysis involved 39 nucleotide sequences each truncated to a length of 292 bp. Here, the green-sulphur bacterium Chlorobium tepidum TLS was chosen as the outgroup.