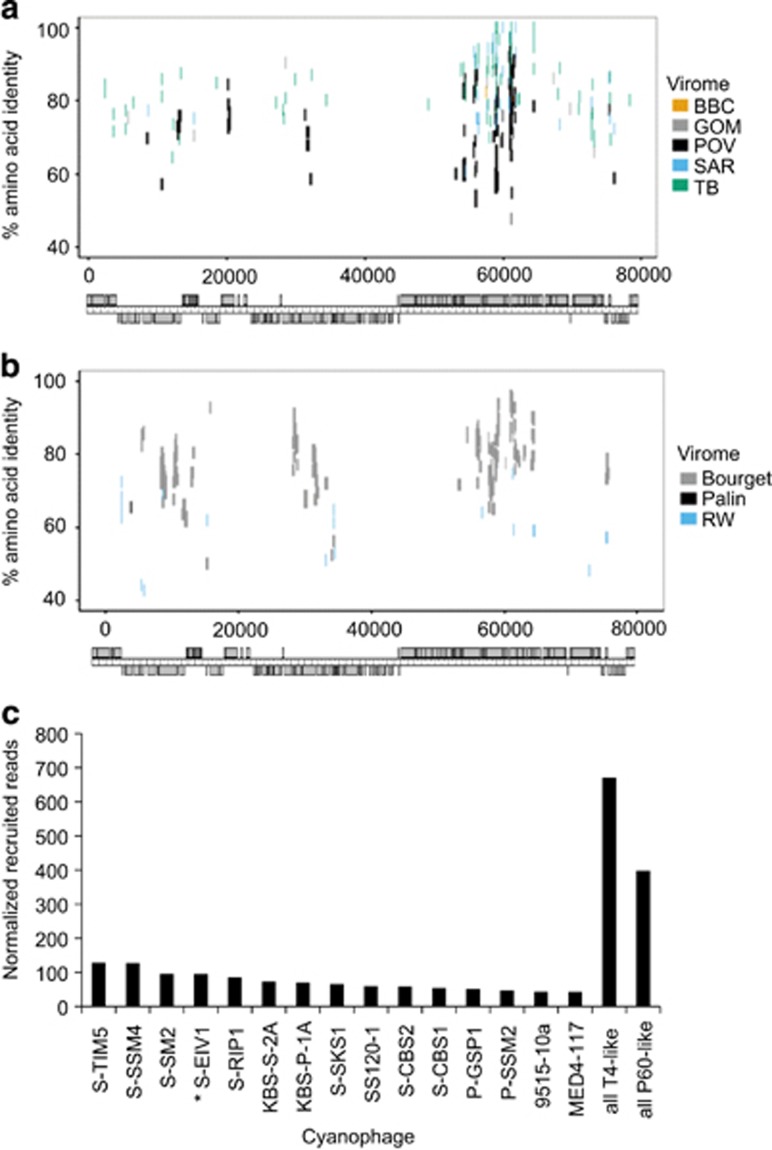
Figure 6.
Prevalence of S-EIV1-like sequences in environmental viral metagenomic data. (a, b) Fragment recruitment of reads from environmental viral metagenomic data (Supplementary Table 2) onto the genome of S-EIV1. Each horizontal line represents a read recruited from one of the following publicly available metagenomic data sets: (a) marine viral metagenomes: Gulf of Mexico (GOM), Strait of Georgia (BBC), Sargasso Sea (SAR), Pacific Ocean (POV) and Tampa Bay (TB). (b) Freshwater viral metagenomes Lake Bourget (Bourget), Lake Pavin (Pavin) and Reclaimed water (RW). Reads were recruited against each of the assembled genomes using tBLASTx with an e-value of 10−10. The position of each line represents the percent similarity of the read to the genome. Only the hit with the highest e-value was used for each read. (c) Abundance of S-EIV1 relative to other cyanophages in the Pacific Ocean Virome. Only the 15 cyanophage isolates that recruited the most reads are shown in the bar chart. The number of reads was normalized to the number of ORFs in each genome.