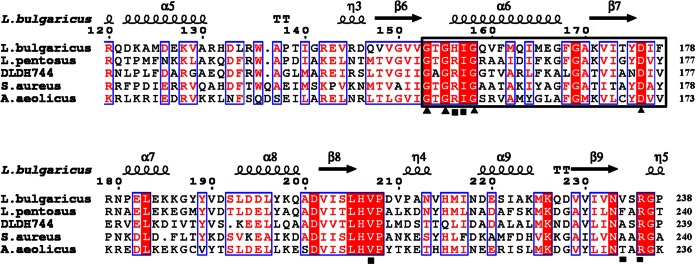
FIG 4.
Sequence alignment of DLDH744 with homologous enzymes. The secondary structure of D-LDH from L. bulgaricus (PDB ID 1J4A) is presented (top). The indicated numbers of the residues refer to the D-LDH from L. bulgaricus. Helices are marked with spirals, beta strands with arrows, and turns with the letter T. Identical residues and conserved substitutions are shaded in red and boxed in by blue rectangles, respectively. The cofactor-binding sites are identified by a black square. The nucleotide-binding signature domain GXGXXG(17X)D is identified by black triangles. Asn174 is a unique residue in DLDH744 that differs from residues in the same position in the other d-2-hydroxyacid dehydrogenases. L. pentosus, Lactobacillus pentosus; S. aureus, Staphylococcus aureus.