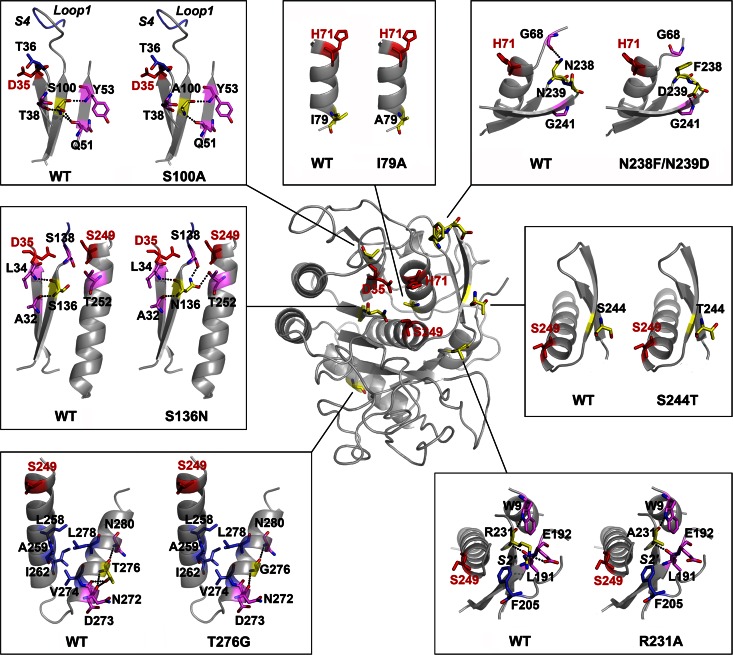
FIG 7.
The effects of substitutions on enzyme structure. An overview of the positions of substituted residues (in yellow) and catalytic triad residues (D35, H71, and Ser249; in red) in the homology model of PBL5X is shown in the center of the picture. Local structures around the original residues in the wild-type (WT) WF146 protease and the substituted residues in PBL5X are shown in boxes. Dotted lines represent hydrogen bonds that are formed by the original and substituted residues in the WT and PBL5X, respectively. The substrate-binding subsites, S4 and S2′, as well as loop 1 of the substrate-binding subsite S4 are indicated in the corresponding boxes.