Abstract
F-specific RNA bacteriophages (FRNAPH) have been widely studied as tools for evaluating fecal or viral pollution in water. It has also been proposed that they can be used to differentiate human from animal fecal contamination. While FRNAPH subgroup I (FRNAPH-I) and FRNAPH-IV are often associated with animal pollution, FRNAPH-II and -III prevail in human wastewater. However, this distribution is not absolute, and variable survival rates in these subgroups lead to misinterpretation of the original distribution. In this context, we studied FRNAPH distribution in urban wastewater and animal feces/wastewater. To increase the specificity, we partially sequenced the genomes of phages of urban and animal origins. The persistence of the genomes and infectivity were also studied, over time in wastewater and during treatment, for each subgroup. FRNAPH-I genome sequences did not show any specific urban or animal clusters to allow development of molecular tools for differentiation. They were the most resistant and as such may be used as fecal or viral indicators. FRNAPH-II's low prevalence and low sequence variability in animal stools, combined with specific clusters formed by urban strains, allowed differentiation between urban and animal pollution by using a specific reverse transcription-PCR (RT-PCR) method. The subgroup's resistance over time was comparable to that of FRNAPH-I, but its surface properties allowed higher elimination rates during activated-sludge treatment. FRNAPH-III's low sequence variability in animal wastewater and specific cluster formation by urban strains also allowed differentiation by using a specific RT-PCR method. Nevertheless, its low resistance restricted it to being used only for recent urban pollution detection. FRNAPH-IV was too rare to be used.
INTRODUCTION
Water contamination by pathogens is a major public health problem. Microorganisms affecting water quality are mainly excreted in feces, and water contamination is caused by discharge of fecal waste of animal origin (e.g., from runoff after rain, farms, and slaughterhouses [SH]) or human origin (e.g., from raw or treated urban wastewater) into the environment. Enteric pathogens include bacteria, protozoa, and enteric viruses (1). The last have been identified in many cases of waterborne and food-borne diseases in developed countries (2).
The virological quality of water is usually indirectly estimated from the number of fecal indicator bacteria, such as Escherichia coli and enterococci (3). However, these indicators have serious limitations, especially for the estimation of viral pollution. First, it has been largely demonstrated that viruses are more resistant than bacteria in the environment; therefore viral outbreaks may still be identified even in the absence of indicator bacteria (4, 5). Second, conventional indicators are present in human and animal feces, and they do not provide information about the origin of fecal contamination (6). Yet, this parameter is essential because the reservoir of many waterborne pathogens, especially viruses, is human (7). Finally, the direct estimation of viral pollution (e.g., noroviruses and hepatitis viruses) by molecular tools could be challenging, not only because of the low concentration in the matrix but also because of the difficulty in interpreting positive results without any information about infectivity. In this context, F-specific RNA bacteriophages (FRNAPH) may be useful tools in a number of situations.
FRNAPH are nonenveloped single-stranded RNA viruses, 20 to 30 nm in diameter, belonging to the Leviviridae family. They are classified in two genera, Levivirus and Allolevivirus, each containing 2 subgroups: FRNAPH-I (MS2-like) and -II (GA-like), which form the genus Levivirus, and FRNAPH-III (Qbeta-like) and -IV (SP-like), which form the genus Allolevivirus (8). These phages reside in the gut of warm-blooded animals and are characterized by their capacity to infect bacteria producing F pili, like E. coli. They get excreted in feces at high frequency and concentrations (9). Considering that they are present in large amounts in wastewater, they may be used as an indicator of viral pollution. These phages can be detected both by rapid culture and by molecular methods (reverse transcription-PCR [RT-PCR]), another advantage compared to pathogenic viruses, which can often only be detected by molecular techniques. FRNAPH are similar in size, shape structure, and genome makeup to the main waterborne pathogenic viruses (e.g., norovirus and hepatitis A virus), and they are often used as surrogates to assess the behavior of enteric viruses in water during treatment (10, 11). Finally, it is assumed that the distribution of the 4 FRNAPH subgroups can be used to differentiate human from animal fecal contamination. Indeed, FRNAPH-I and -IV are generally associated with animal pollution, whereas FRNAPH-II and -III make up the majority in human wastewater (12, 13). For this reason, subgroup distribution has been widely studied to determine the origin of fecal contamination in river water (14, 15), shellfish (16, 17), and sediments (18).
However, there are two main limits to the use of these indicators to track the origin of fecal pollution. First of all, the distribution of phage subgroups is not absolute. (i) FRNAPH-III, generally associated with humans, has been isolated in high proportions in swine wastewater (12, 19), (ii) FRNAPH-I has been detected in municipal wastewater (20), and (iii) even if FRNAPH-II seems to be highly associated with humans (21), it has also been found in animal feces (22). The second limitation is due to the variable survival rates of FRNAPH in the environment. Indeed, FRNAPH-I and -II are less affected by environmental stresses (23–26), thereby being often found in surface waters, regardless of the source of pollution (27–29). As a consequence, this may cause misinterpretation of the subgroup distribution when estimated by infectious-phage isolation.
The aim of this work was to evaluate the potential of FRNAPH as a tool to differentiate human from animal fecal pollution. We explored in particular the possibility of minimizing both drawbacks described above. The first objective was to determine whether genome sequencing within each subgroup could allow better differentiation between animal and human fecal contamination as recently suggested (20, 30). To achieve this, we isolated by culture FRNAPH strains from samples of known origin (animal feces, urban wastewater, and slaughterhouse wastewater). The strains were subjected to genotyping, and the maturation protein gene was sequenced with the aim to design more-specific RT-PCR methods for human and animal phages. The second objective was to determine whether direct genome detection by RT-PCR could reduce the bias caused by differences in the survival rates of each subgroup in the environment. For this purpose, we compared the persistence of infectious phages with that of the genomes of each subgroup over time and assessed the impact of water treatment on these parameters.
MATERIALS AND METHODS
Animal feces samples.
Forty-seven fecal samples were collected from different areas in northwestern France from May to October 2014. The samples were stool from cattle (n = 10), swine (n = 8), sheep (n = 9), horses (n = 10), chickens (n = 5), and ducks/geese (n = 5). All samples were collected in sterile polypropylene containers and kept at −20°C during transport.
After thawing, 4 g of stool was mixed with 28 ml of phosphate-buffered saline (PBS) for 3 min in a DT-50 tube with Ultra-Turrax tube drive (IKA-Werke GmbH & Co. KG, Staufen, Germany). Three milliliters of the mixture (stool concentration of 0.125 g/ml) was removed for RNA extraction and treated as described below. A volume of 14.5 ml of PBS–0.3% peptone was added to obtain a final concentration of 0.1% peptone (final stool concentration of 0.083 g/ml). The suspension was mixed for 1 min and kept in ice for 3 h. Finally, 6 ml of chloroform was added to 20 ml of the stool suspension, and after centrifugation (2,000 × g for 5 min) the supernatant was collected for culture.
Slaughterhouse samples.
Four wastewater samples were collected from a cattle slaughterhouse (CattleSH-1 to -4) and from a swine slaughterhouse (SwineSH-1 to -4) located in eastern France. The first site generates a wastewater flow of 450 m3/day when slaughtering 250 animals. The second site processes 30 to 90 swine per day. Wastewater samples were collected in sterile glass bottles and kept at 4°C for less than 3 h prior to analysis.
Urban sewage samples.
Five urban sewage samples were collected at the entry of a wastewater treatment plant (WWTP-1 to -5) receiving 80,000 m3/day from an urban area in eastern France, with a population of approximately 260,000. All samples were collected in sterile glass bottles and kept at 4°C for less than 3 h prior to analysis. Another sample was collected from a smaller WWTP located in northwestern France (WWTP-6), receiving wastewater from 8,500 inhabitants. The sample was kept at −20°C during transport.
Finally, in order to isolate the most resistant phages, 2 samples (WWTP-4 and WWTP-5) were stored at 4°C in the dark, in glass bottles, and analyzed over time. Two effluent samples were also collected from the first WWTP, whose processes include pretreatment, primary treatment by sedimentation, secondary treatment by activated sludge, and tertiary treatment for dephosphorylation.
Infectious FRNAPH enumeration and phage plaque isolation.
FRNAPH were detected and enumerated by the double-agar-layer technique according to International Organization for Standardization (ISO) standard 10705-1 (31). Salmonella enterica serovar Typhimurium WG49 (NCTC 12484) was used as the host strain (32), and kanamycin and nalidixic acid were added to obtain a final antibiotic concentration of 100 μg/ml. Culture was performed directly from 1 ml of several dilutions of wastewater and stool suspensions in 90-mm-diameter petri dishes. For each dilution, 2 assays were performed. Viral concentration was expressed in PFU per milliliter after an 18-h incubation period. For low-concentration samples (WWTP effluents or aged samples), enumeration was performed twice from 5 ml of wastewater in 150-mm-diameter petri dishes to lower the detection limit to 0.1 PFU/ml. Several phage plaques were isolated from the positive samples, collected with tips, and suspended in 1 ml of PBS–5% glycerol. After brief vortexing, the suspension was filtered through sterile Acrodisc syringe filters (pore size, 0.22 μm; Pall Life Sciences, Ann Arbor, MI). RNA was then extracted as described below.
RNA extraction.
To detect genomes in the samples, extraction was performed directly from 1 ml of wastewater (WWTP or SH). For the stool samples, 3 ml of the suspension prepared as previously described (stool concentration of 0.125 g/ml) was mixed with 1 ml of chloroform and centrifuged (2,000 × g for 5 min). Extraction was then performed from 1 ml of the supernatant. It was carried out by adding 3 ml of Isol-RNA lysis reagent (5 PRIME GmbH, Hilden, Germany) to the samples, followed by gentle agitation for 10 min on a tube roller at 70 rpm (Starlab International GmbH, Hamburg, Germany). After addition of 1 ml of chloroform, the mixture was stirred vigorously by hand for 15 s and centrifuged (2,000 × g for 2 min) in 15 ml of Phase Lock Gel Heavy (5 PRIME GmbH, Hilden, Germany). RNA was extracted from the supernatant using NucliSens magnetic extraction reagents (bioMérieux, Marcy l'Etoile, France) on NucliSens MiniMAG according to the manufacturer's recommendations. RNA was concentrated in 100 μl of elution buffer and stored at −80°C in 1.5-ml DNA LoBind tubes (Eppendorf, Hamburg, Germany).
RNA from phage plaques was extracted using NucliSens EasyMag (bioMérieux, Marcy l'Etoile, France) from 50 μl of phage suspension, eluted in 100 μl of buffer, and stored at −80°C.
Real-time RT-PCR.
For the direct detection of genomes, RNA suspensions were subjected to the RT-PCR method proposed by Wolf et al. (22) with some modifications. Separate reactions were carried out for each subgroup. Reverse transcription was performed using SuperScript III reverse transcriptase (Life Technologies, Carlsbad, CA) according to the manufacturer's recommendations from 7.5 μl of RNA using 20 pmol of reverse primer, 100 U of SuperScript III, and 20 U of RNasin (Promega, Madison, WI) in a 20-μl reaction volume. PCR was then carried out from 5 μl of cDNA with TaqMan universal PCR master mix (Life Technologies, Carlsbad, CA) according to the manufacturer's recommendations in a 25-μl reaction volume, with reverse and forward primers at a concentration of 1 μM and the probe at a concentration of 0.3 μM. PCR amplification was performed at 50°C for 2 min and 95°C for 10 min, followed by 45 cycles of 15 s at 95°C and 1 min at 60°C on a StepOne Plus real-time PCR system (Life Technologies, Carlsbad, CA).
Infectious phage genotyping was performed using phage plaque RNA extracted by a 1-step multiplex quantitative RT-PCR using the primers and probes designed by Wolf et al. (22). The reaction was performed with the RNA UltraSense one-step quantitative RT-PCR system (Life Technologies, Carlsbad, CA) according to the manufacturer's recommendations from 2 μl of RNA with 20 U of RNasin (Promega, Madison, WI), each primer at a concentration of 0.3 μM, and the probes at a concentration of 0.15 μM in 20-μl reaction volume. The reaction was carried out on a StepOne Plus real-time PCR system (Life Technologies, Carlsbad, CA) at 55°C for 60 min (reverse transcription) and 95°C for 5 min (hot start), followed by 40 cycles of 15 s at 95°C (denaturation) and 40 s at 58°C (annealing/extension). The primers and probes proposed by Ogorzaly and Gantzer (33) were also tested for the detection of some phage strains under the same conditions.
FRNAPH sequencing.
Amplification was performed with a OneStep RT-PCR kit (Qiagen, Hilden, Germany) on RNA extracted from phage plaques, according to the manufacturer's recommendations. For each subgroup, primers were designed based on consensus sequences retrieved in the GenBank database, National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/GenBank/index.html). Alignments and consensus sequence determination were performed using Seaview software v.4 (34), and designs were performed with Primer3Plus software (35). Percent sequence similarity was calculated using the ClustalW2 program of the European Bioinformatics Institute (EBI) of the EMBL (http://www.ebi.ac.uk/Tools/msa/clustalw2/).
For FRNAPH-I and -II, the amplified fragment was about 1,300 to 1,400 nucleotides in length, including the entire maturation protein gene (Table 1). For FRNAPH-III, the size of the amplified fragment located on the maturation protein gene was about 400 nucleotides. The amplified RT-PCR products were purified and sequenced by a sequencing service provider (Beckman Coulter Genomics, Takeley, United Kingdom). Nucleotide sequences were aligned using Seaview software v.4 (34), and phylogenetic trees were generated using BioNJ analysis, with distance estimations based on the Kimura two-parameter equation (K2P). Confidence values of branches were calculated by bootstrap analysis using 100 replications. Designs of trees were then performed using Figtree software v.1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/).
TABLE 1.
Primer sequences used for sequencing reactions
| Target virus | Primer sequence (5′–3′) | Sense | Position | Product length (bp) |
|---|---|---|---|---|
| FRNAPH-I | GGG GTC CTG CTC AAC TTC CT | + | 17–36b | 1,407 |
| ACC CCG TTA GCG AAG TTG CTa | − | 1404–1423b | ||
| FRNAPH-II | CAT GCC GTT AGG TTT AGN TGA | + | 89–109c | 1,280 |
| GTA CCG CCA TTA TCG ACG AG | − | 1349–1368c | ||
| FRNAPH-III | GTG TCC GAY TGG AAG GAR CT | + | 941–960d | 405 |
| CAT GAT CDA ATT GAC CCA AWG | − | 1325–1345d |
Primer selected from a previous study (33).
Genome location of primers based on GenBank accession number NC_001417.2, MS2 phage.
Genome location of primers based on GenBank accession number NC_001426.1, GA phage.
Genome location of primers based on GenBank accession number NC_001890.1, Qbeta phage.
Corresponding sequences of FRNAPH available in GenBank were added to the trees. Strains and accession numbers for FRNAPH-I are as follows: 253A, HQ332484.1; 862A, HQ332489.1; DL1, EF107159.1; DL16, EF108464.1; DL52, JQ966307.1; DL54, JQ966308.1; JP501, AF227251.1; L1.G10.6, FJ799473.1; L2.G10.13, FJ799546.1; L3.G40.5, FJ799645.1; M12, AF195778.1; MS2, NC_001417.2; and ST4, EF204940.1. Those for FRNAPH-II are as follows: DL10, FJ483837.1; DL20, FJ483839.1; GA, NC_001426.1; KU1, AF227250.1; and T72, FJ483838.1. Those for FRNAPH-III are as follows: 2010JunDWTP11-13, AB627184.1; 1FR, JF719735.1; 2FR, JF719736.1; 3FR, JF719737.1; AncP1, AB971354.1; BR12, FJ483842.1; BZ1, FJ483844.1; HL4-9, FJ483841.1; MX1, S81548.1; M11, S81550.1; Qbeta, NC_001890.1; Qbeta_3, GQ153930.1; TW18, FJ483840.1; and VK, FJ483843.1.
Statistical analysis.
Analysis of covariance (ANCOVA) was used to test the difference in linearity of each genome and infectious-phage persistence. Statistical analyses were performed using StatEL (AD Science, Paris, France). Differences were considered significant when P was <0.05.
RESULTS
FRNAPH in stool samples.
FRNAPH concentration was evaluated after culture by plaque assay and direct genome detection in animal feces. Samples from 7 different species (n = 47) were analyzed (Table 2). Analysis of individual stool specimens revealed variable incidences of infectious phages depending on the species. Plaque formation was detected in a total of 9 (19%) fecal samples: no FRNAPH were isolated from cattle feces, but at least 1 sample for the other species was positive. Except for cattle samples, prevalence in fecal specimens was between 10% and 40%, with the highest prevalence in duck/goose and chicken specimens. Concentrations of infectious FRNAPH in stool samples ranged from 1.2 × 101 to 1.9 × 103 PFU/g for swine, 1.6 × 104 to 1.9 × 104 PFU/g for sheep, and 5.1 × 101 to 3.0 × 102 PFU/g for ducks/geese. The highest concentrations were found in chicken manure, with a maximum of 1.1 × 105 PFU/g.
TABLE 2.
Incidence and concentration of infectious FRNAPH and genomes in feces and wastewater
| Sample source | Infectivity |
Total genome |
||
|---|---|---|---|---|
| No. of positive samples/no. taken | Concn rangea | No. of positive samples/no. taken | Concn rangea | |
| Feces | ||||
| Cattle | 0/10 | 2/10 | 3.9–4.1 | |
| Swine | 2/8 | 1.1–3.3 | 5/8 | 3.8–5.8 |
| Sheep | 2/9 | 4.2–4.3 | 5/9 | 3.7–6.5 |
| Horses | 1/10 | 1.2 | 1/10 | 4.8 |
| Chickens | 2/5 | 3.3–5.0 | 3/5 | 3.7–8.9 |
| Ducks and geese | 2/5 | 1.7–2.5 | 3/5 | 4.0–5.4 |
| Wastewater | ||||
| Urban WWTP | 6/6 | 2.3–4.2 | 6/6 | 4.9–5.8 |
| CattleSH | 4/4 | 4.7–5.2 | 4/4 | 6.4–6.8 |
| SwineSH | 4/4 | 2.4–4.5 | 4/4 | 4.7–6.1 |
Concentrations are expressed in log10 PFU/g of stool or log10 PFU/ml of wastewater.
FRNAPH genomes were more often detected in stool samples than infectious phages. Depending on the species, 10% to 62.5% of the samples were positive, with the highest prevalence in swine feces (Table 2). Ten samples contained genomes, while no infectious phages were found (specimens from 2 cattle, 3 swine, 3 sheep, 1 chicken, and 1 duck). The concentration of genomes was higher than that of infectious phages, with a ratio of between 1.1 and 3.9 log10. The average concentration for each species ranged from 1.0 × 104 genome copies (gc)/g (cattle) to 2.7 × 108 gc/g (chickens), with the highest concentrations observed in samples with the largest amounts of infectious FRNAPH.
FRNAPH in wastewater.
FRNAPH concentration was also evaluated by plaque assay and direct genome detection in urban wastewater and in cattle slaughterhouse (CattleSH) and swine slaughterhouse (SwineSH) wastewater. All 14 samples were positive for infectious phage (Table 2). The average concentration was 5.7 × 103 PFU/ml in urban wastewater, with the lowest concentration in the sample from the small WWTP located in northwestern France. In CattleSH waters, the average concentration was 1.1 × 105 PFU/ml, and it was about 8.6 × 103 PFU/ml in SwineSH waters.
In all the samples, genomes were found in higher concentrations than infectious phages, with ratios of between 1.5 and 2.6 log10. The average concentration was 3.3 × 105 gc/ml in urban wastewater, 4.3 × 106 gc/ml in CattleSH wastewater, and 5.6 × 105 gc/ml in SwineSH wastewater.
FRNAPH subgroup distribution.
Distribution was determined directly in the samples by detection of subgroup genomes and after culture by phage plaque genotyping. Phage plaque distribution in feces and wastewater is shown in Table 3. Among the 376 phage isolates using S. Typhimurium WG49 as host (32), 91.2% gave a positive RT-PCR signal for a subgroup (n = 343).
TABLE 3.
Distribution of infectious FRNAPH subgroups isolated from feces and wastewater
| Sample source | No. of plaques picked | No. of plaques classified as: |
||||
|---|---|---|---|---|---|---|
| FRNAPH | FRNAPH-I | FRNAPH-II | FRNAPH-III | FRNAPH-IV | ||
| Feces | ||||||
| Cattle | 0 | |||||
| Swine | 14 | 13 | 12 | 1 | ||
| Sheep | 20 | 20 | 20 | |||
| Horses | 3 | 1 | 1 | |||
| Chickens | 24 | 24 | 24 | |||
| Ducks and geese | 25 | 24 | 13 | 11 | ||
| Wastewater | ||||||
| Urban WWTP | 115 | 106 | 28 | 77 | 1 | |
| CattleSH | 117 | 103 | 100 | 3 | ||
| SwineSH | 58 | 52 | 40 | 12 | ||
| Total | 376 | 343 | 110 | 39 | 177 | 17 |
The 14 plaques tested from swine feces were typed as 86% (n = 12) formed by FRNAPH-I and 7% (n = 1) by FRNAPH-IV. As regards sheep and chicken stool specimens, 20 and 24 plaques, respectively, were picked and all were formed by FRNAPH-I. Only 3 phage plaques were isolated from horse feces; 1 was formed by FRNAPH-I, but the other two did not give positive signals for any subgroup. In the 2 duck fecal samples, 25 phage plaques were typed; the first specimen contained only FRNAPH-I (n = 8) and 1 uncharacterized plaque, while the second specimen contained 31% (n = 5) FRNAPH-I plaques and 69% (n = 11) FRNAPH-II plaques.
One hundred fifteen phages were isolated from the urban wastewater samples. Of the 92% of plaques that were characterized (n = 106), 26% were formed by FRNAPH-II (n = 28) and 72% by FRNAPH-III (n = 77). Among the 117 phages isolated from CattleSH waters, 88% gave positive RT-PCR signals (n = 103), among which 100 were FRNAPH-III and 3 FRNAPH-IV. Finally, among the phages isolated from SwineSH waters (n = 58), 40 were FRNAPH-I and 12 FRNAPH-IV. All FRNAPH-IV phages were isolated from the same specimen, representing 92% of the phages characterized in this sample.
This distribution was confirmed overall by direct detection of genomes in the samples. Most genomes detected in the stool samples were that of FRNAPH-I, though with some exceptions: 100% of the genomes detected in 1 sheep stool sample were that of FRNAPH-III (1.0 × 105 gc/g), but this sample was negative for infectious phages. The FRNAPH-II genome was detected in the duck fecal sample in which were found infectious FRNAPH-II (2.2 × 105 gc/g, representing 93% of the total genome).
The genomes detected in the urban wastewater samples were mostly those of FRNAPH-II (6.0 × 104 to 2.2 × 105 gc/ml, representing 30% to 61% of the total number of genomes) and those of FRNAPH-III (2.9 × 104 to 4.7 × 105 gc/ml, representing 30% to 67% of the total number of genomes). FRNAPH-I and -IV genomes were detected at lower concentrations (respectively, 7.8 × 103 and 4.1 × 103 gc/ml on average), representing less than 6% of all the genomes detected. With regard to SwineSH wastewater, the FRNAPH-I genome was detected in all the samples (5.1 × 104 to 1.3 × 106 gc/ml). The FRNAPH-IV genome was detected in 1 sample, representing 6% of the total genomes detected in this specimen. Finally, in all CattleSH wastewater samples, the FRNAPH-III genome prevailed (>99%), with an average concentration of 4.3 × 106 gc/ml. The FRNAPH-I genome was found in 3 samples (1.1 × 103 to 1.8 × 104 gc/ml), the FRNAPH-II genome was found in 2 samples (1.5 × 103 to 2.0 × 103 gc/ml), and the FRNAPH-IV genome was found in 1 sample (9.2 × 103 gc/ml).
The presence and isolation of FRNAPH-II and -III strains of animal origin (ducks and cattle) seem particularly interesting for evaluating the potential of FRNAPH to track the origin of fecal pollution. These subgroups, generally associated with human pollution, were sequenced and compared with strains isolated from urban wastewater. Conversely, no FRNAPH-I strains were isolated from the raw WWTP samples.
Genome and infectious FRNAPH stability in urban wastewater.
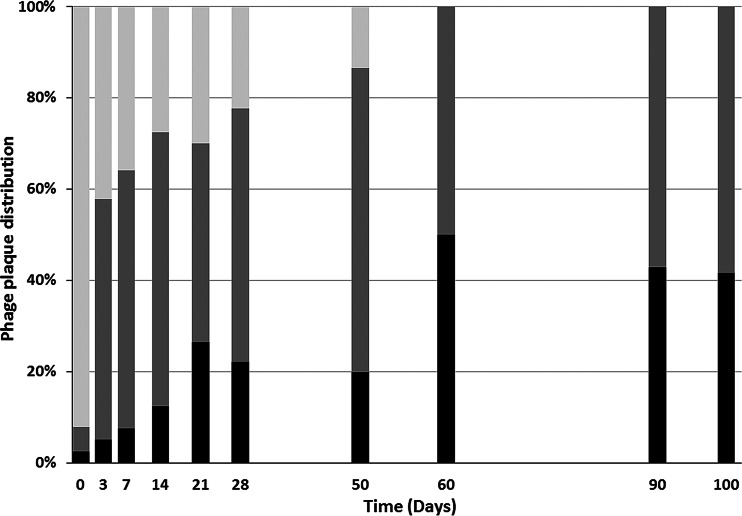
The stability of FRNAPH (genomes and infectious phages) was evaluated in wastewater over time and after wastewater treatment. Two raw sewage samples (WWTP-4 and WWTP-5) were analyzed over time. On the first day, infectious FRNAPH concentrations were, respectively, 3.8 × 103 and 8.0 × 103 PFU/ml. Infectious FRNAPH subgroup distribution over time is shown in Fig. 1. Initially, FRNAPH-III prevailed (>90%) in raw wastewater. Its proportion decreased steadily over time and was less than 50% after 3 days. The FRNAPH-II proportion was hidden during the first days by the large number of FRNAPH-III phages, but it was between one- and two-thirds of the total number of infectious FRNAPH after 3 days. Finally, the proportion of FRNAPH-I phages was low during the first days, but it increased steadily to one-half of the infectious phages after 60 days.
FIG 1.
Infectious FRNAPH-I (black), FRNAPH-II (dark gray), and FRNAPH-III (light gray) distribution in urban wastewater. Between 16 and 24 plaques were isolated and typed for each analysis.
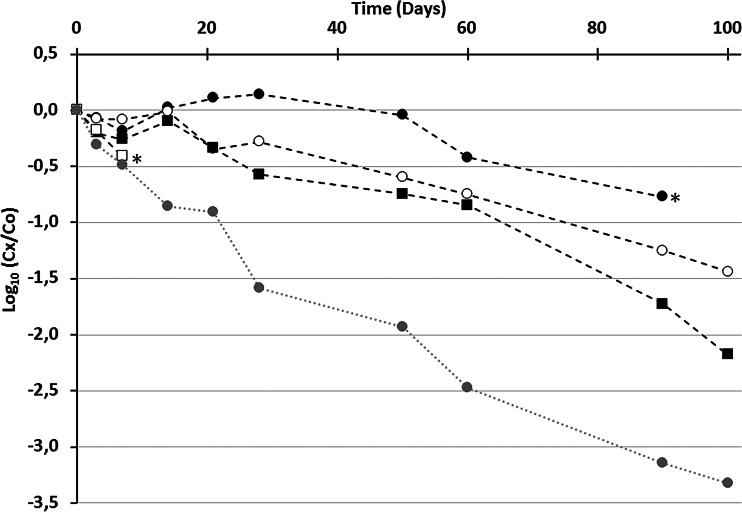
While infectivity decreased significantly (time required for 90% loss of infectivity [T90] estimated at day 21.7), genomes were overall more persistent over time (Fig. 2). The FRNAPH-IV genome seemed to be the least resistant, but the rapid achievement of quantification limits due to low concentrations in the original samples prevented a trend from being accurately determined. The genome of FRNAPH-I turned out to be the most resistant under these conditions (decrease of <1 log10 after 100 days), and a significant but low difference between its persistence and that of FRNAPH-II was observed (P value = 0.027, ANCOVA). No marked difference between the persistence of the FNRAPH-II genome and that of FRNAPH-III was observed during the first 60 days (P value = 0.69, ANCOVA). The genomic search approach thus appears more reliable to estimate subgroup distribution, especially in the case of an aged fecal contamination, thereby reducing the survival bias.
FIG 2.
Persistence of FRNAPH-I (black circles), FRNAPH-II (white circles), FRNAPH-III (black squares), and FRNAPH-IV (white squares) genomes, as well as of infectious FRNAPH (gray circles) in urban wastewater kept at 4°C in the dark. *, quantification limit; Cx, concentration at the indicated time; Co, initial concentration.
Urban WWTP effluents (n = 2) were analyzed to estimate the impact of water treatment on infectious FRNAPH and genome concentration. The average concentration of infectious FRNAPH in effluents was found to be 33.2 PFU/ml. Among the plaques typed (n = 28), 86% were formed by FRNAPH-I and the rest by FRNAPH-II. The genome of FRNAPH-I was detected at concentrations up to 1.1 × 103 gc/ml. Genomes of other subgroups were not detected, mainly because of the low volume analyzed. During wastewater treatment, the infectivity decreased by 2.3 log10 units and the genome copies decreased by 1.0 log10 unit for FRNAPH-I. As FRNAPH-II and -III genome concentrations in effluents were below the detection limit, losses during water treatment were, respectively, at least 2.0 log10 and 1.9 log10.
The change in the FRNAPH subgroup distribution over time in urban wastewater and the analysis of effluents allowed FRNAPH-I strains of urban origin to be isolated; these were compared with animal strains by sequencing.
FRNAPH sequencing.
Sequencing was performed on FRNAPH isolated by culture from samples of different origins. A two-phase strategy was used. First, we sequenced a representative panel of the supposed animal subgroup (FRNAPH-I) isolated from animal samples and the supposed human subgroups (FRNAPH-II and -III) isolated from urban wastewater. Then, we sequenced most FRNAPH-II and -III strains isolated from animal samples on the one hand and most FRNAPH-I strains isolated from urban samples on the other hand. Altogether, we sequenced the entire maturation protein genes of 158 FRNAPH-I and 70 FRNAPH-II strains, as well as part of those of 92 FRNAPH-III strains.
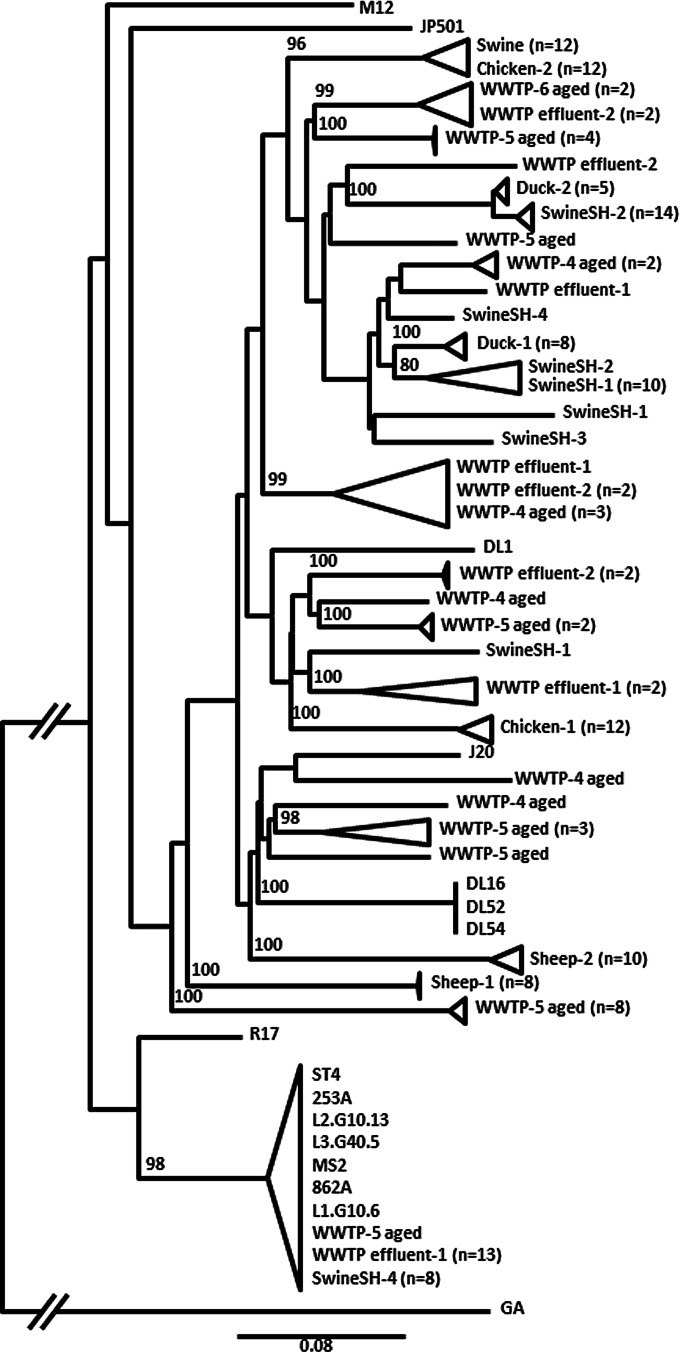
FRNAPH-I strains of animal origin were isolated from feces or slaughterhouse (SH) waters. Group formation during sequence comparison appeared to be sample dependent, and each stool sample formed a robust grouping (Fig. 3). In each stool sample, FRNAPH-I sequences presented a high degree of similarity (>98.6%). However, no relationship between sequences isolated from different animals of the same species was observed. For example, the percentage of identity between the 2 sheep samples was only 90.7% ± 0.3%; it was 92.8% ± 0.4% between the 2 chicken samples, and it was 94.5% ± 0.3% between the 2 duck samples. FRNAPH-I strains isolated from swine feces were not grouped with any strains from SwineSH wastewater samples, but they were close to isolates from the Chicken-2 sample (>98.7% similarity). More variability was observed in SwineSH wastewater samples (93.2% similarity in SwineSH-2 and only 90.9% in SwineSH-4). Groupings were again sample dependent, except for 1 strain isolate from SwineSH-2, which was close to the group formed by strains from SwineSH-1 (98.0% ± 0.2% similarity).
FIG 3.
Phylogenetic relationship of FRNAPH-I strains isolated from urban WWTPs and animal slaughterhouse (SH) wastewater or fecal samples based on the maturation protein gene (1,182 nucleotides). Distances were determined by using the K2P model, and the tree was plotted by the BioNJ method. FRNAPH-II GA (NC_001426.1) was used as an outgroup. Only bootstrap values >70% are shown.
FRNAPH-I strains of urban origin were isolated from WWTP samples. They were present in low density in urban wastewater but showed high resistance; therefore urban strains could be isolated from aged samples or effluents. Sequences were spread out over the entire phylogenetic tree obtained with animal FRNAPH-I strains. No relationship appeared between them, and some isolates were close to strains of animal origin. For example, many strains isolated from urban WWTP effluent 1 were close to strains from SwineSH-4, and all grouped with prototype phage MS2. In this cluster, percent sequence variation was only 0.7%.
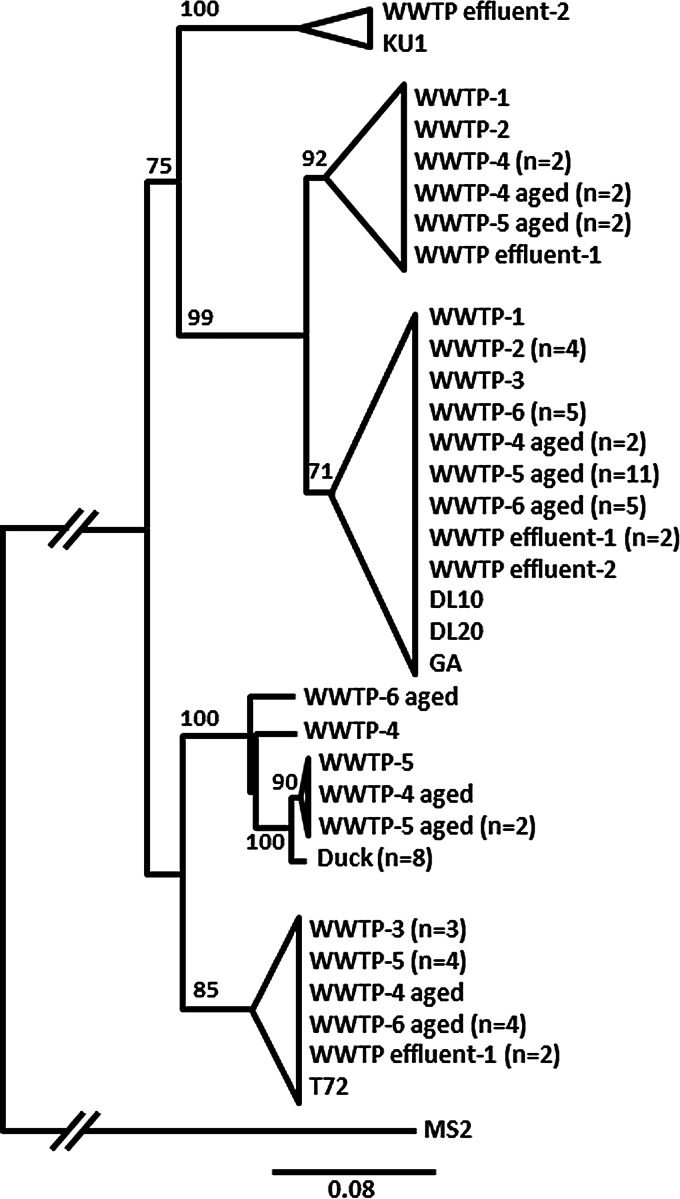
FRNAPH-II strains of human origin were isolated from urban wastewater. Percent sequence identity between them ranged from 82.3% to 100%. The isolated strains could be grouped into several clusters (Fig. 4). No sample-dependent grouping was observed, and the FRNAPH isolated from WWTP-6 (urban WWTP located in another geographical area) were present in all clusters.
FIG 4.
Phylogenetic relationship of FRNAPH-II strains isolated from urban WWTPs and a duck fecal sample based on the maturation protein gene (1,173 nucleotides). Distances were determined by using the K2P model, and the tree was plotted by the BioNJ method. FRNAPH-I MS2 (NC_001417.2) was used as an outgroup. Only bootstrap values >70% are shown.
All FRNAPH-II strains isolated from animals were sequenced. They all originated from a duck fecal sample and were grouped into a cluster with strains coming from urban wastewater. Even if urban strains and duck strains were close, with a maximum similarity of 98.7% in this cluster, FRNAPH-II strains from the duck sample formed a separate grouping because all isolates exhibited identical sequences. All previously mentioned FRNAPH-II strains were detected by using multiplex RT-PCR as proposed by Wolf et al. (22), but the singularity of the duck strains was confirmed by testing the primer and probe design described by Ogorzaly and Gantzer (33). This PCR assay targeted the 3′ end of the replicase gene and allowed most urban FRNAPH-II strains to be detected, leaving out strains isolated from duck feces. More precisely, out of the 6 urban strains belonging to the cluster formed with the duck strains, half gave a typical positive RT-PCR signal, while the others were not detected. Out of the 10 strains tested from the T72 cluster, 70% gave a typical positive signal, 1 gave a late signal, and 2 were uncharacterized. Finally, out of the 10 strains tested from other clusters, 100% were detected.
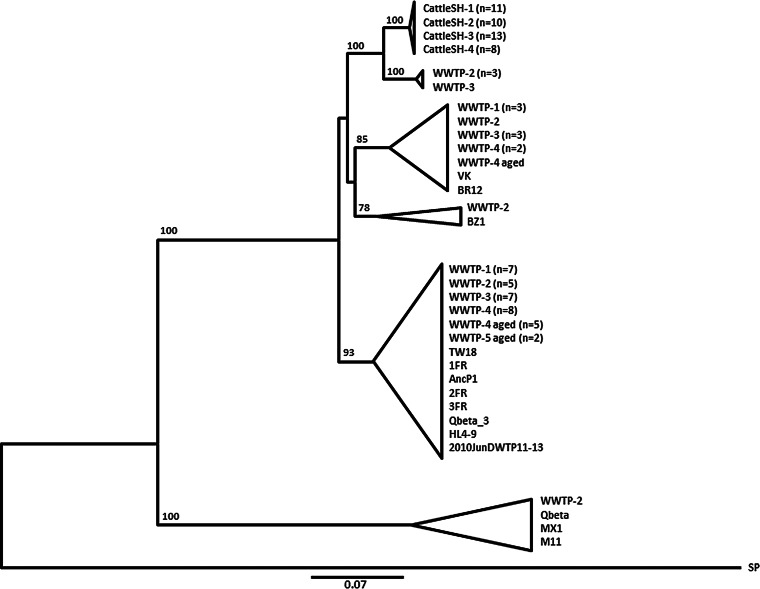
FRNAPH-III strains of human origin were isolated from urban wastewater. Percent sequence identity between them was 85.3% ± 14.7%. They were grouped into several clusters (Fig. 5), often associated with one or more known strains available in GenBank. One cluster was formed by a group composed of a large number of isolates (n = 34), with most strains already published (TW18, 1FR, AncP1, 2FR, 3FR, Qbeta_3, HL4-9, and 2010JunDWTP11-13). The degree of similarity ranged between 90.0% and 100% for these phages.
FIG 5.
Phylogenetic relationship of FRNAPH-III strains isolated from urban WWTP and cattle slaughterhouse (CattleSH) wastewater based on partial nucleotide sequences of the maturation protein gene (380 nucleotides). Distances were determined by using the K2P model, and the tree was plotted by the BioNJ method. FRNAPH-IV SP (X07489.1) was used as an outgroup. Clusters with bootstrap values >70% are collapsed.
Most FRNPAH-III strains originating from animals were also sequenced. They were all isolated from CattleSH wastewater. These strains (n = 42) formed a robust cluster with low sequence variations (0% to 1.3%), corresponding to a maximum variation of 5 nucleotides. Among them, 15 isolates had identical sequences and were present in all samples. Similarly, 2 other clone sequences were identified, one in 12 isolates and the other in 7 isolates. These were present in 3 of the 4 samples.
A group formed by 4 FRNAPH strains from the urban WWTP samples was close to the cattle cluster. Percent sequence identity with cattle strains was 94.2% for the most genetically distant ones and 95.8% for the closest ones, but the bootstrap value (100) confirmed the strength of the cattle cluster. As observed for FRNAPH-II, all FRNAPH-III strains were detected by using the multiplex RT-PCR designed by Wolf et al. (22), but the genetic difference between cattle strains was confirmed by the RT-PCR method proposed by Ogorzaly and Gantzer (33) targeting a coat protein gene region. Using the latter, no FRNAPH strains isolated from the CattleSH samples were characterized, whereas all those tested (n = 24), isolated from the urban WWTP samples, including the 4 strains forming the cluster close to the CattleSH group, gave a positive signal for subgroup III.
DISCUSSION
The present work aimed to study F-specific RNA phages (FRNAPH) in stool or wastewater and, in particular, their use as tracers to differentiate urban from animal fecal contamination.
FRNAPH have been used for years as fecal pollution indicators in the environment (36, 37) because of their high prevalence and concentration in stool or wastewater. Our results confirmed this application because FRNAPH were found in very high concentrations in all animal or urban wastewater. Concentrations ranged between 2.3 and 5.2 log10 PFU/ml in wastewater when using the double-agar-layer technique. In comparison, other authors detected infectious FRNAPH concentrations of 2.9 to 4.2 log10 PFU/ml in urban wastewater (15, 38–40), while Schaper et al. (13) observed concentrations ranging from 3.4 to 4.6 log10 PFU/ml in slaughterhouse wastewater. Genome concentrations ranged between 4.7 and 6.8 log10 gc/ml. This type of analysis has been less frequently described in the literature; Wolf et al. (22) observed an average concentration of 3.5 log10 gc/ml in raw urban wastewater, while Hata et al. (40) and Flannery et al. (41) detected only about 2.0 log10 gc/ml. In these last two studies, genomes were detected in lower concentration than infectious FRNAPH, which suggests an underestimation of genome concentration. In our samples, the ratio of genomes to infectious phages ranged from 1.5 to 2.6 log10.
The prevalence in animal stool samples was found to be low as previously described (12, 42), but, in the case of positivity, concentrations were significant. They ranged between 1.1 and 5.0 log10 PFU/g for infectious FRNAPH and between 3.7 and 8.9 log10 gc/g for genomes. To sum up, the FRNAPH concentrations obtained in this study are very similar to those reported in the literature.
The main goal of the present work was to investigate the potential of FRNAPH to differentiate between urban and animal pollution. Genotyping before or after culture using the RT-PCR method designed by Wolf et al. (22) clearly confirmed the predominance of FRNAPH-I in stool or wastewater of animal origin and the predominance of FRNAPH-II and -III in raw urban wastewater, as described in other studies (12, 13, 15, 43, 44). Unfortunately, our results also underlined that FRNAPH genotyping is not sufficient to determine the origin of fecal contamination, since FRNAPH-II and -III were also detected in animal samples. In the same way, the FRNAPH-I genome was detected in urban wastewater. Further knowledge on the variability of sequences within each subgroup according to their animal or urban origin is therefore clearly needed to use FRNAPH as an indicator of the origin of fecal pollution. This was our first objective. We chose to perform sequencing on the maturation protein gene because it shows a high degree of variability (45).
No common point between FRNAPH-I sequences from urban wastewater and those of animal origin was observed. The urban FRNAPH-I strains may have a real human origin, or they may come from variety of animals present in the urban area, but in smaller numbers than the 260,000 inhabitants. In any event, detection of FRNAPH-I by RT-PCR targeting the maturation protein gene does not allow distinguishing urban from animal fecal contamination in the environment. FRNAPH-II was never detected in slaughterhouse wastewater either by direct genomic search or after culture. Nevertheless, 11 FRNAPH-II sequences were isolated in a single duck stool sample. All sequences were identical. In addition, if the primers and probe proposed by Wolf et al. (22) were able to detect the duck isolates, the approach proposed by Ogorzaly and Gantzer (33) was not. Indeed, it appears that, in our experiment, the latter RT-PCR allowed specific detection of urban wastewater FNRAPH-II isolates, even if some isolates were missed.
FRNAPH-III strains were more frequently detected in animal wastewater. Forty-two strains were isolated from cattleSH wastewater using RT-PCR as described by Wolf et al. (22). Again, all these sequences clearly clustered together and did not show any overlapping with the urban strains. The RT-PCR method developed by Ogorzaly and Gantzer (33) did not allow detection of these isolates, while it allowed that of the urban strains. The narrow cluster formed by FRNAPH-III isolated from the cattle slaughterhouse and the presence of identical sequences in the samples collected over more than 3 months (December 2013 to March 2014) suggested a specific signature of this slaughterhouse over time. Based on the 5′ end of the maturation protein gene, Stewart et al. (30) also observed genetic variations among FRNAPH-III strains isolated from urban WWTPs and from swine lagoons, with specific strains from each swine lagoon.
Practically speaking, the RT-PCR assays designed by Ogorzaly and Gantzer (33) seem more specific for urban FRNAPH-II and -III than those designed by Wolf et al. (22). Such a high degree of specificity is explained by the use of minor groove binder (MGB) probes (46). These results show the bias related to the choice of primers/probes and their spectral detection, but such a choice may be of benefit depending on the strategy pursued. If detection of all FRNAPH is required, the probes designed by Wolf et al. (22) may be more suitable as they provide better sensitivity (18). Their recent design based on all sequences available in GenBank in 2010 may confer the largest spectrum of detection, while Ogorzaly's and Gantzer's FRNAPH-II and -III probes (33) may be preferable for microbial source tracking.
If specificity is an important point, persistence of FRNAPH in the environment is another one. It has been clearly demonstrated in the literature that FRNAPH subgroups have different survival rates in the environment. Their behavior has already been studied using prototypic phages. Yang and Griffiths (24) observed better persistence of infectious MS2 (FRNAPH-I) and GA (FRNAPH-II) than of Qbeta (FRNAPH-III) under various conditions. Muniesa et al. (23) reached the same conclusion by studying naturally occurring infectious FRNAPH in environmental waters (in situ conditions). Nevertheless, levels of persistence of the genomes of the subgroups have never been compared in natural waters. Since it is clearly admitted that the viral genome is more resistant than infectious (47–51), our second objective was to study the inactivation rate of infectious FRNAPH and the persistence of each subgroup genome. Two situations were explored: the persistence over time in raw wastewater at 4°C and the impact of a standard wastewater treatment plant. The parameters used during persistence experiments were set to limit the growth of bacteria and yeasts in samples and to be the most favorable for a long survival of FRNAPH.
Even if the genome of FRNAPH-I fell just above the significance threshold as being more resistant than those of FRNAPH-II and -III, on the whole, genomes were more persistent than infectious, with a loss of less than 1.0 log10 after 60 days, versus 2.5 log10 for infectivity. Infectious FRNAPH-I and -II strains had the best survival rates, with a ratio of 1:1 in wastewater after 60 days. Infectious FRNAPH-III strains were the least resistant because, although they were in the majority in raw wastewater (>90%), they were in the minority after 3 days and they were no longer detected after 50 days of incubation.
During water treatment (activated-sludge process), infectious FRNAPH concentration was reduced by 2.4 log10. In WWTPs with similar water treatment systems, De Luca et al. (52) and Hata et al. (40) observed a decrease in the number of infectious FRNAPH of 3 log10, while Lucena et al. (53) noted a reduction of 2 log10 in winter and 3 log10 in summer. In effluents, infectious FNRAPH-I prevailed, with the presence of a few FRNAPH-II strains, but only the FRNAPH-I genome was detectable. Given that the persistence of FRNAPH-I is close to that of FRNAPH-II, it may be supposed that FRNAPH-II strains were physically eliminated during the process. Other studies demonstrated that infectious FRNAPH-I strains showed higher resistance to water treatment using activated sludge, while FRNAPH-II strains were strongly adsorbed on activated sludge but remained infectious (15, 54), which may be explained by different surface properties depending on the subgroup (55).
In our study, the high prevalence of FRNAPH-III before treatment as opposed to that of FRNAPH-I after treatment indicates that the reduction observed was mainly due to FRNAPH-III disappearance. We may hypothesize that the more FRNAPH-III strains there are in raw water, the higher the estimated removal rate will be, and the more FRNAPH-I strains there are, the lower the removal rate will be. For this reason, it may be interesting to perform genotyping before studying removal treatment when FRNAPH strains are used as surrogates.
To conclude, this study has brought forward important considerations when using FRNAPH as a tool to differentiate urban from animal fecal contamination. FRNAPH-I genome sequences did not show any specific clusters of urban or animal origin and thus cannot allow molecular tools to be designed for differentiation. However, as they were the most resistant in wastewater and during water treatment, they may be used as fecal or viral indicators. FRNAPH-II's low prevalence and low sequence variability in animal stool, combined with specific clusters formed by urban strains, allows it to be related to urban pollution by using specific RT-PCR such as the one developed by Ogorzaly and Gantzer (33). Its resistance over time was comparable to that of FRNAPH-I, but its surface properties may have allowed more elimination during activated-sludge treatment, requiring the use of sensitive methods to detect it in the environment. FRNAPH-III's low sequence variability in animal wastewater, combined with specific clusters formed by urban strains, also allows differentiation between urban and animal pollution by using the RT-PCR approach of Ogorzaly and Gantzer (33). However, its very low resistance over time or during water treatment restricts its use to recent fecal contaminations by raw wastewater. FRNAPH-IV was too rare to be used.
ACKNOWLEDGMENTS
The work has been partly funded by Agence de l'Eau Seine Normandie (AESN), Zone Atelier du bassin de la Moselle (ZAbM), and Institut Carnot Energie et Environnement en Lorraine (ICEEL). It has also been supported by Unité Mixte Technologique ViroControl.
We thank Labeo Manche for the supply of samples.
REFERENCES
- 1.Ashbolt NJ. 2004. Microbial contamination of drinking water and disease outcomes in developing regions. Toxicology 198:229–238. doi: 10.1016/j.tox.2004.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Field KG, Samadpour M. 2007. Fecal source tracking, the indicator paradigm, and managing water quality. Water Res 41:3517–3538. doi: 10.1016/j.watres.2007.06.056. [DOI] [PubMed] [Google Scholar]
- 3.Tallon P, Magajna B, Lofranco C, Leung KT. 2005. Microbial indicators of faecal contamination in water: a current perspective. Water Air Soil Pollut 166:139–166. doi: 10.1007/s11270-005-7905-4. [DOI] [Google Scholar]
- 4.Chalmers JWT, McMillan JH. 1995. An outbreak of viral gastroenteritis associated with adequately prepared oysters. Epidemiol Infect 115:163–167. doi: 10.1017/S0950268800058222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DePaola A, Jones JL, Woods J, Burkhardt W III, Calci KR, Krantz JA, Bowers JC, Kasturi K, Byars RH, Jacobs E, Williams-Hill D, Nabe K. 2010. Bacterial and viral pathogens in live oysters: 2007 United States market survey. Appl Environ Microbiol 76:2754–2768. doi: 10.1128/AEM.02590-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gordon DM, Cowling A. 2003. The distribution and genetic structure of Escherichia coli in Australian vertebrates: host and geographic effects. Microbiology 149:3575–3586. doi: 10.1099/mic.0.26486-0. [DOI] [PubMed] [Google Scholar]
- 7.Soller JA, Schoen ME, Bartrand T, Ravenscroft JE, Ashbolt NJ. 2010. Estimated human health risks from exposure to recreational waters impacted by human and non-human sources of faecal contamination. Water Res 44:4674–4691. doi: 10.1016/j.watres.2010.06.049. [DOI] [PubMed] [Google Scholar]
- 8.Bollback JP, Huelsenbeck JP. 2001. Phylogeny, genome evolution, and host specificity of single-stranded RNA bacteriophage (family Leviviridae). J Mol Evol 52:117–128. doi: 10.1007/s002390010140. [DOI] [PubMed] [Google Scholar]
- 9.Grabow WOK. 2001. Bacteriophages: update on application as models for viruses in water. Water SA 27:251–268. doi: 10.4314/wsa.v27i2.4999. [DOI] [Google Scholar]
- 10.Marti E, Monclús H, Jofre J, Rodriguez-Roda I, Comas J, Balcázar JL. 2011. Removal of microbial indicators from municipal wastewater by a membrane bioreactor (MBR). Bioresour Technol 102:5004–5009. doi: 10.1016/j.biortech.2011.01.068. [DOI] [PubMed] [Google Scholar]
- 11.Jebri S, Hmaied F, Jofre J, Mariem Y, Mendez J, Barkallah I, Hamdi M. 2013. Effect of gamma irradiation on bacteriophages used as viral indicators. Water Res 47:3673–3678. doi: 10.1016/j.watres.2013.04.036. [DOI] [PubMed] [Google Scholar]
- 12.Cole D, Long SC, Sobsey MD. 2003. Evaluation of F+ RNA and DNA coliphages as source-specific indicators of fecal contamination in surface waters. Appl Environ Microbiol 69:6507–6514. doi: 10.1128/AEM.69.11.6507-6514.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schaper M, Jofre J, Uys M, Grabow WOK. 2002. Distribution of genotypes of F-specific RNA bacteriophages in human and non-human sources of faecal pollution in South Africa and Spain. J Appl Microbiol 92:657–667. doi: 10.1046/j.1365-2672.2002.01600.x. [DOI] [PubMed] [Google Scholar]
- 14.Ogorzaly L, Tissier A, Bertrand I, Maul A, Gantzer C. 2009. Relationship between F-specific RNA phage genogroups, faecal pollution indicators and human adenoviruses in river water. Water Res 43:1257–1264. doi: 10.1016/j.watres.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 15.Haramoto E, Otagiri M, Morita H, Kitajima M. 2012. Genogroup distribution of F-specific coliphages in wastewater and river water in the Kofu basin in Japan. Lett Appl Microbiol 54:367–373. doi: 10.1111/j.1472-765X.2012.03221.x. [DOI] [PubMed] [Google Scholar]
- 16.Mieszkin S, Caprais MP, Le Mennec C, Le Goff M, Edge TA, Gourmelon M. 2013. Identification of the origin of faecal contamination in estuarine oysters using Bacteroidales and F-specific RNA bacteriophage markers. J Appl Microbiol 115:897–907. doi: 10.1111/jam.12260. [DOI] [PubMed] [Google Scholar]
- 17.Wolf S, Hewitt J, Rivera-Aban M, Greening GE. 2008. Detection and characterization of F+ RNA bacteriophages in water and shellfish: application of a multiplex real-time reverse transcription PCR. J Virol Methods 149:123–128. doi: 10.1016/j.jviromet.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 18.Haramoto E, Kitajima M, Katayama H, Asami M, Akiba M, Kunikane S. 2009. Application of real-time PCR assays to genotyping of F-specific phages in river water and sediments in Japan. Water Res 43:3759–3764. doi: 10.1016/j.watres.2009.05.043. [DOI] [PubMed] [Google Scholar]
- 19.Sundram A, Jumanlal N, Ehlers M. 2007. Genotyping of F-RNA coliphages isolated from wastewater and river water samples. Water SA 32:65–70. doi: 10.4314/wsa.v32i1.5241. [DOI] [Google Scholar]
- 20.Lee JE, Lim MY, Kim SY, Lee S, Lee H, Oh HM, Hur HG, Ko G. 2009. Molecular characterization of bacteriophages for microbial source tracking in Korea. Appl Environ Microbiol 75:7107–7114. doi: 10.1128/AEM.00464-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harwood VJ, Boehm AB, Sassoubre LM, Vijayavel K, Stewart JR, Fong TT, Caprais MP, Converse RR, Diston D, Ebdon J, Fuhrman JA, Gourmelon M, Gentry-Shields J, Griffith JF, Kashian DR, Noble RT, Taylor H, Wicki M. 2013. Performance of viruses and bacteriophages for fecal source determination in a multi-laboratory, comparative study. Water Res 47:6929–6943. doi: 10.1016/j.watres.2013.04.064. [DOI] [PubMed] [Google Scholar]
- 22.Wolf S, Hewitt J, Greening GE. 2010. Viral multiplex quantitative PCR assays for tracking sources of fecal contamination. Appl Environ Microbiol 76:1388–1394. doi: 10.1128/AEM.02249-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muniesa M, Payan A, Moce-Llivina L, Blanch AR, Jofre J. 2009. Differential persistence of F-specific RNA phage subgroups hinders their use as single tracers for faecal source tracking in surface water. Water Res 43:1559–1564. doi: 10.1016/j.watres.2008.12.038. [DOI] [PubMed] [Google Scholar]
- 24.Yang Y, Griffiths MW. 2013. Comparative persistence of subgroups of F-specific RNA phages in river water. Appl Environ Microbiol 79:4564–4567. doi: 10.1128/AEM.00612-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Long SC, Sobsey MD. 2004. A comparison of the survival of F+RNA and F+DNA coliphages in lake water microcosms. J Water Health 2:15–22. [PubMed] [Google Scholar]
- 26.Schaper M, Duran AE, Jofre J. 2002. Comparative resistance of phage isolates of four genotypes of F-specific RNA bacteriophages to various inactivation processes. Appl Environ Microbiol 68:3702–3707. doi: 10.1128/AEM.68.8.3702-3707.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stewart-Pullaro J, Daugomah JW, Chestnut DE, Graves DA, Sobsey MD, Scott GI. 2006. F+ RNA coliphage typing for microbial source tracking in surface waters. J Appl Microbiol 101:1015–1026. doi: 10.1111/j.1365-2672.2006.03011.x. [DOI] [PubMed] [Google Scholar]
- 28.Brion GM, Meschke JS, Sobsey MD. 2002. F-specific RNA coliphages: occurrence, types, and survival in natural waters. Water Res 36:2419–2425. doi: 10.1016/S0043-1354(01)00547-4. [DOI] [PubMed] [Google Scholar]
- 29.Griffin DW, Stokes R, Rose JB, Paul JH III. 2000. RNA coliphage assay to identify fecal sources in Homosassa. Microb Ecol 39:56–64. doi: 10.1007/s002489900193. [DOI] [PubMed] [Google Scholar]
- 30.Stewart JR, Vinje J, Oudejans SJG, Scott GI, Sobsey MD. 2006. Sequence variation among group III F-specific RNA coliphages from water samples and swine lagoons. Appl Environ Microbiol 72:1226–1230. doi: 10.1128/AEM.72.2.1226-1230.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.International Organization for Standardization. 2001. ISO 10705-1. Water quality: detection and enumeration of bacteriophages. Part 1: enumeration of F-specific RNA bacteriophages. International Organization for Standardization, Geneva, Switzerland. [Google Scholar]
- 32.Havelaar AH, Hogeboom WM. 1984. A method for the enumeration of male-specific bacteriophages in sewage. J Appl Bacteriol 56:439–447. doi: 10.1111/j.1365-2672.1984.tb01372.x. [DOI] [PubMed] [Google Scholar]
- 33.Ogorzaly L, Gantzer C. 2006. Development of real-time RT-PCR methods for specific detection of F-specific RNA bacteriophage genogroups: application to urban raw wastewater. J Virol Methods 138:131–139. doi: 10.1016/j.jviromet.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 34.Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 35.Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JAM. 2007. Primer3Plus, an enhanced Web interface to Primer3. Nucleic Acids Res 35:W71–W74. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yahya M, Hmaied F, Jebri S, Jofre J, Hamdi M. 2015. Bacteriophages as indicators of human and animal fecal contamination in raw and treated wastewaters from Tunisia. J Appl Microbiol 118:1217–1225. doi: 10.1111/jam.12774. [DOI] [PubMed] [Google Scholar]
- 37.Lucena F, Ribas F, Duran AE, Skraber S, Gantzer C, Campos C, Morón A, Calderón E, Jofre J. 2006. Occurrence of bacterial indicators and bacteriophages infecting enteric bacteria in groundwater in different geographical areas. J Appl Microbiol 101:96–102. doi: 10.1111/j.1365-2672.2006.02907.x. [DOI] [PubMed] [Google Scholar]
- 38.Lucena F, Méndez X, Morón A, Calderón E, Campos C, Guerrero A, Cárdenas M, Gantzer C, Shwartzbrood L, Skraber S, Jofre J. 2003. Occurrence and densities of bacteriophages proposed as indicators and bacterial indicators in river waters from Europe and South America. J Appl Microbiol 94:808–815. doi: 10.1046/j.1365-2672.2003.01812.x. [DOI] [PubMed] [Google Scholar]
- 39.Contreras-Coll N, Lucena F, Mooijman K, Havelaar A, Pierz V, Boque M, Gawler A, Höller C, Lambiri M, Mirolo G, Moreno B, Niemi M, Sommer R, Valentin B, Wiedenmann A, Young V, Jofre J. 2002. Occurrence and levels of indicator bacteriophages in bathing waters throughout Europe. Water Res 36:4963–4974. doi: 10.1016/S0043-1354(02)00229-4. [DOI] [PubMed] [Google Scholar]
- 40.Hata A, Kitajima M, Katayama H. 2013. Occurrence and reduction of human viruses, F-specific RNA coliphage genogroups and microbial indicators at a full-scale wastewater treatment plant in Japan. J Appl Microbiol 114:545–554. doi: 10.1111/jam.12051. [DOI] [PubMed] [Google Scholar]
- 41.Flannery J, Keaveney S, Rajko-Nenow P, O'Flaherty V, Doré W. 2013. Norovirus and FRNA bacteriophage determined by RT-qPCR and infectious FRNA bacteriophage in wastewater and oysters. Water Res 47:5222–5231. doi: 10.1016/j.watres.2013.06.008. [DOI] [PubMed] [Google Scholar]
- 42.Mauffret A, Caprais MP, Gourmelon M. 2012. Relevance of Bacteroidales and F-specific RNA bacteriophages for efficient fecal contamination tracking at the level of a catchment in France. Appl Environ Microbiol 78:5143–5152. doi: 10.1128/AEM.00315-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blanch AR, Belanche-Muñoz L, Bonjoch X, Ebdon J, Gantzer C, Lucena F, Ottoson J, Kourtis C, Iversen A, Kühn I, Mocé L, Muniesa M, Schwartzbrod J, Skraber S, Papageorgiou GT, Taylor H, Wallis J, Jofre J. 2006. Integrated analysis of established and novel microbial and chemical methods for microbial source tracking. Appl Environ Microbiol 72:5915–5926. doi: 10.1128/AEM.02453-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Noble RT, Allen SM, Blackwood AD, Chu W, Jiang SC, Lovelace GL, Sobsey MD, Stewart JR, Wait DA. 2003. Use of viral pathogens and indicators to differentiate between human and non-human fecal contamination in a microbial source tracking comparison study. J Water Health 1:195–207. [PubMed] [Google Scholar]
- 45.Friedman SD, Genthner FJ, Gentry J, Sobsey MD, Vinjé J. 2009. Gene mapping and phylogenetic analysis of the complete genome from 30 single-stranded RNA male-specific coliphages (family Leviviridae). J Virol 83:11233–11243. doi: 10.1128/JVI.01308-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yao Y, Nellåker C, Karlsson H. 2006. Evaluation of minor groove binding probe and Taqman probe PCR assays: influence of mismatches and template complexity on quantification. Mol Cell Probes 20:311–316. doi: 10.1016/j.mcp.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 47.Gassilloud B, Schwartzbrod L, Gantzer C. 2003. Presence of viral genomes in mineral water: a sufficient condition to assume infectious risk? Appl Environ Microbiol 69:3965–3969. doi: 10.1128/AEM.69.7.3965-3969.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skraber S, Gassilloud B, Schwartzbrod L, Gantzer C. 2004. Survival of infectious poliovirus-1 in river water compared to the persistence of somatic coliphages, thermotolerant coliforms and poliovirus-1 genome. Water Res 38:2927–2933. doi: 10.1016/j.watres.2004.03.041. [DOI] [PubMed] [Google Scholar]
- 49.Fong TT, Lipp EK. 2005. Enteric viruses of humans and animals in aquatic environments: health risks, detection, and potential water quality assessment tools. Microbiol Mol Biol Rev 69:357–371. doi: 10.1128/MMBR.69.2.357-371.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choi S, Jiang SC. 2005. Real-time PCR quantification of human adenoviruses in urban rivers indicates genome prevalence but low infectivity. Appl Environ Microbiol 71:7426–7433. doi: 10.1128/AEM.71.11.7426-7433.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ogorzaly L, Bertrand I, Paris M, Maul A, Gantzer C. 2010. Occurrence, survival, and persistence of human adenoviruses and F-specific RNA phages in raw groundwater. Appl Environ Microbiol 76:8019–8025. doi: 10.1128/AEM.00917-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Luca G, Sacchetti R, Leoni E, Zanetti F. 2013. Removal of indicator bacteriophages from municipal wastewater by a full-scale membrane bioreactor and a conventional activated sludge process: implications to water reuse. Bioresour Technol 129:526–531. doi: 10.1016/j.biortech.2012.11.113. [DOI] [PubMed] [Google Scholar]
- 53.Lucena F, Duran AE, Morón A, Calderón E, Campos C, Gantzer C, Skraber S, Jofre J. 2004. Reduction of bacterial indicators and bacteriophages infecting faecal bacteria in primary and secondary wastewater treatments. J Appl Microbiol 97:1069–1076. doi: 10.1111/j.1365-2672.2004.02397.x. [DOI] [PubMed] [Google Scholar]
- 54.Haramoto E, Fujino S, Otagiri M. 2015. Distinct behaviors of infectious F-specific RNA coliphage genogroups at a wastewater treatment plant. Sci Total Environ 520:32–38. doi: 10.1016/j.scitotenv.2015.03.034. [DOI] [PubMed] [Google Scholar]
- 55.Dika C, Ly-Chatain MH, Francius G, Duval JFL, Gantzer C. 2013. Non-DLVO adhesion of F-specific RNA bacteriophages to abiotic surfaces: importance of surface roughness, hydrophobic and electrostatic interactions. Colloids Surfaces A Physicochem Eng Asp 435:178–187. doi: 10.1016/j.colsurfa.2013.02.045. [DOI] [Google Scholar]