Figure 1.
Pilot Screen for Known Regulators of Endosome-to-Golgi Retrieval Using Anti-CD8 Uptake Assay
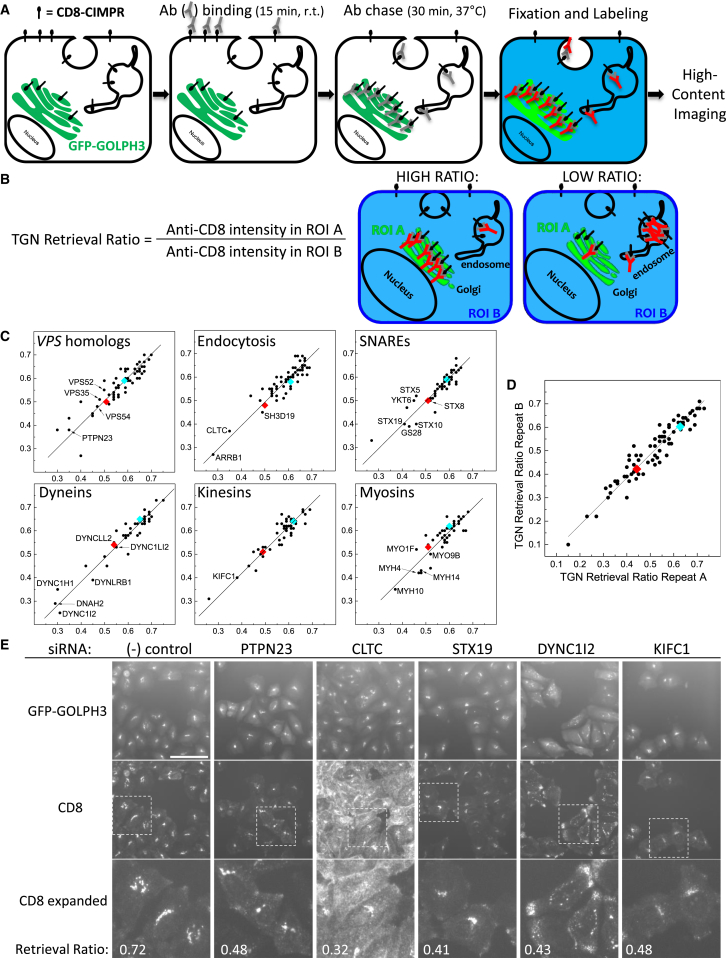
(A) Schematic of the anti-CD8 antibody retrieval assay as adapted for high-throughput screening. HeLa cells stably expressing a CD8-CIMPR reporter and the Golgi protein GFP-GOLPH3 were used. Anti-CD8 antibody (Ab) was bound at room temperature for 15 min and chased at 37°C for 30 min. Labeling and imaging details are in Supplemental Experimental Procedures.
(B) Definition of the TGN retrieval ratio used in our studies and depiction of subcellular antibody localizations that give rise to high or low TGN retrieval ratios.
(C) Scatterplots of the TGN retrieval ratios measured in replicate screens of 60 VPS gene homologs, 60 endocytosis genes, 43 SNARE protein genes, 41 dynein genes, 49 kinesin genes, and 43 myosin genes. Negative control measurements are in blue; SNX1 siRNA positive control measurements are in red. A linear fit to the data points is also shown. Marked data points indicate known endosome-to-Golgi retrieval pathway genes or genes that were selected for further validation.
(D) Replicate TGN retrieval ratios for the validation screen of the pilot study, in which 80 individual ON TARGETplus siRNA sequences (four per gene) were assayed. Controls and linear fit are as in (C).
(E) Representative images of the pilot validation screen for the negative control (no siRNA) and for five sets of siRNA-treated cells with reduced TGN retrieval ratio. Different phenotypes that give rise to reduced TGN retrieval ratios are discussed in the text. Scale bar, 100 μm (top two rows) and 38 μm (bottom row).