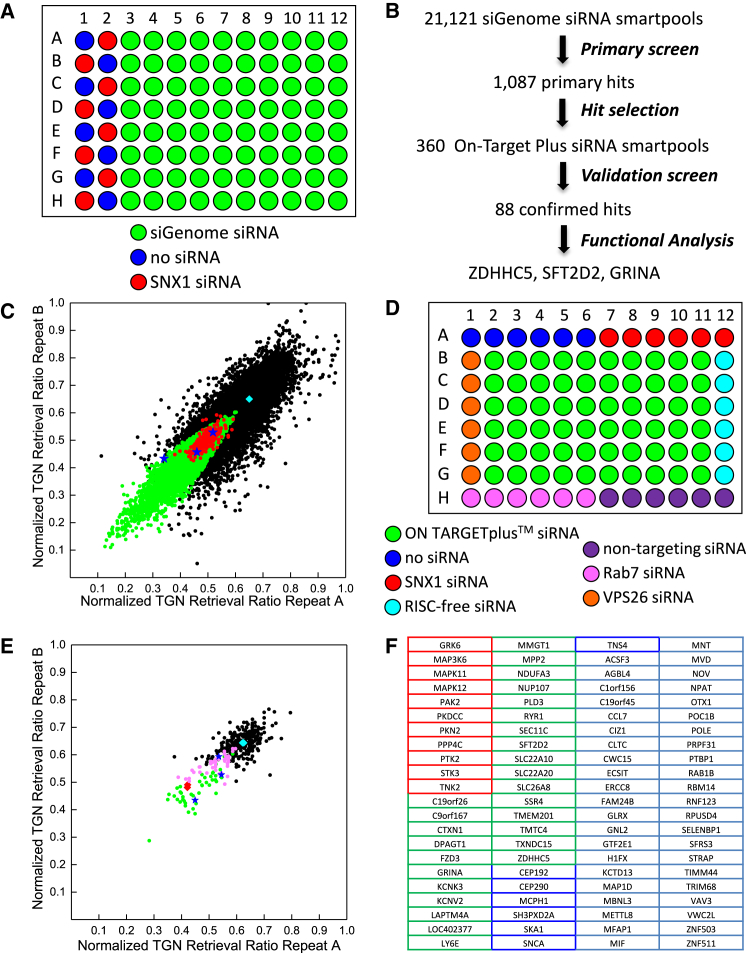
Figure 2.
A Whole-Genome siRNA Screen to Identify Regulators of Endosome-to-Golgi Retrieval
(A) Plate layout for genome-wide loss-of-function screen.
(B) Whole-genome screen hit selection.
(C) Scatterplot of the normalized TGN retrieval ratios for the two replicates of the whole-genome screen. Each data point represents a single siRNA pool. Only valid measurements (18,465 siRNA pools) are included. Negative (cyan square) and positive (SNX1 siRNA, red data points) controls are shown. Green data points indicate siRNA pools with TGN retrieval ratio SSMD ≤ (−3) (very strong hits). Blue star data points indicate the three hits characterized further in Figures 3, 4, 5, and 6.
(D) Plate layout for the ON TARGETplus validation screen.
(E) Replicate normalized TGN retrieval ratios for the ON TARGETplus smartpool validation screen (360 pools). The colors used are as in (C), with additional magenta data points indicating siRNA pools with TGN retrieval ratio SSMD between (−2) and (−3) (strong hits).
(F) Table listing the 88 genes that were very strong or strong hits in the validation screen. Red-bordered cells indicate kinase and phosphatase genes, green-bordered cells contain genes encoding membrane proteins, and blue-bordered cells contain genes encoding cytoskeletal proteins. More information about these genes is provided in Table S4.