Figure 3.
Characterization of SFT2D2 Function in Endosome-to-Golgi Retrieval
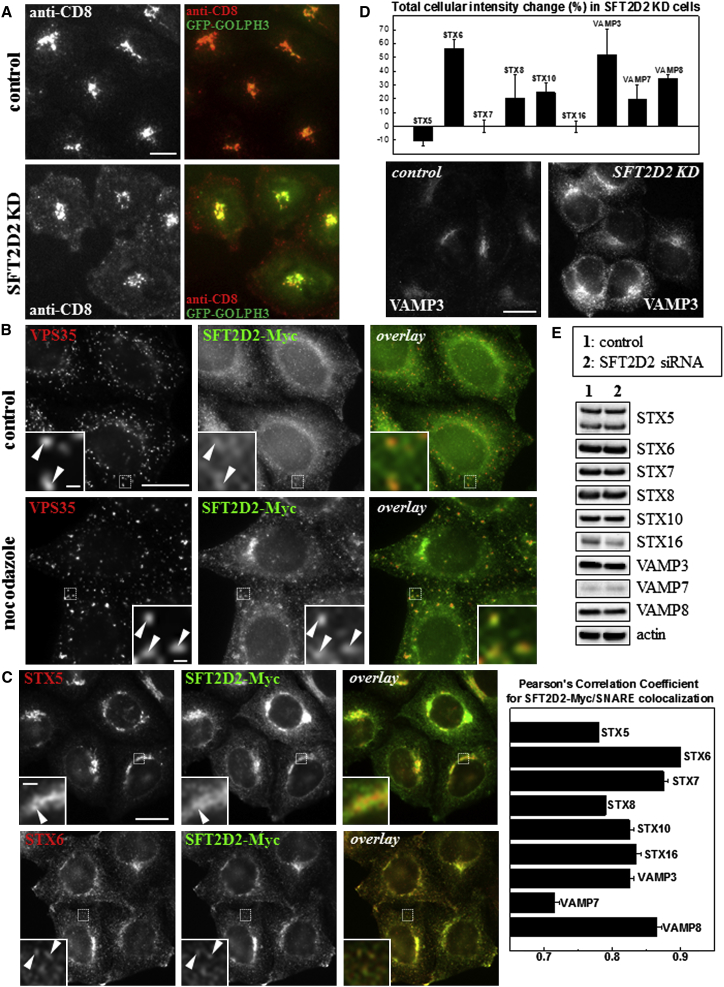
(A) Primary screen anti-CD8 antibody-uptake images for control cells (top) and SFT2D2 KD cells (bottom).
(B) Cells stably expressing SFT2D2-Myc show localization of SFT2D2 to perinuclear membranes and endosomes, including VPS35-positive endosomes (arrowheads in inset). Treatment with nocodazole disperses the endosomes and even more clearly shows colocalization of SFT2D2-Myc and VPS35 (arrowheads in inset).
(C) Cells stably expressing SFT2D2-Myc were costained with antibodies against various post-Golgi SNARE proteins. Colocalization was quantified by Pearson’s correlation coefficient (right-hand graph) and indicates very extensive colocalization with STX6, STX7, and VAMP8. The image panels illustrate colocalization of SFT2D2-Myc and STX5 at the Golgi (top, arrowhead) but much more extensive colocalization of SFT2D2-Myc and STX6 (bottom, arrowheads). In (B) and (C), the white dashed box delineates the area magnified in the insets.
(D) SNARE protein staining was compared for control and SFT2D2 KD HeLa cells and quantified. The graph shows the change in cellular intensity measured for each post-Golgi SNARE investigated. Images illustrate the increased cellular intensity of VAMP3 in SFT2D2 KD cells.
(E) Control and SFT2D2 KD HeLa cell lysates were separated by LDS-PAGE and blotted for the indicated SNARE proteins or actin. Scale bars in (A)–(D), 20 μm, except insets in (B), 1 μm, and in (C), 2 μm. Quantitation in (C) and (D) was done using automated microscopy (see Experimental Procedures); error bars indicate SD of two separate multicell experiments. Average Pearson’s correlation coefficients measured were often identical in repeated experiments.