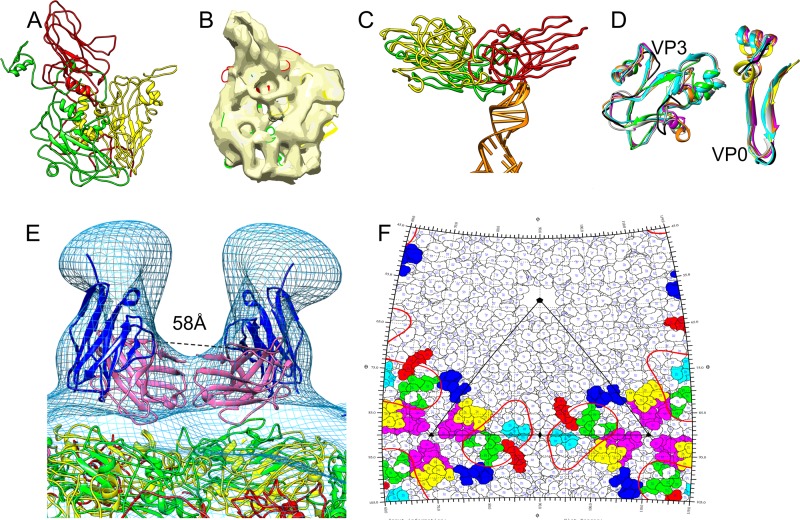
FIG 5.
Epitopes on HPeV1 for AM28. (A) Homology models of VP1 (red), VP0 (yellow), and VP3 (green) built using I-TASSER. (B) Final fits of VP1, VP0, and VP3 homology models into an asymmetric unit of HPeV1 (EMD-1690). (C) One of the capsid protein interactions with RNA in HPeV1 is shown. An RNA stem-loop structure (PDB ID 3P22, chain A; orange) was rigidly fitted into the previously described (6) finger-like density in the HPeV1 EM density map (EMD-1690). The majority of the RNA interactions appear to be with VP1 and VP3. (D) Mapping the epitope region (black) on other picornaviruses by superimposing asymmetric units of echovirus 1 (PDB ID 1EV1; magenta), poliovirus 1 (PDB ID 1POV; gray), enterovirus 71 (PDB ID 3VBF; orange), and foot-and-mouth disease virus (PDB ID 1QQP; cyan) on final fits of HPeV1 VP0 (yellow) and VP3 (green). For ease of visualization, only the regions encompassing the epitopes in VP0 and VP3 are shown. (E) Mapping epitopes for AM28 on HPeV1 surface by fitting the AM28 Fab variable region homology model into the Fab density seen in HPeV1-AM28 Fab reconstruction and superimposing VP1 (red), VP0 (yellow), and VP3 (green) fits for HPeV1 (EMD-1690) into the HPeV1-AM28-Fab reconstruction (mesh). AM28 variable heavy chain is shown in magenta, and variable light chain is shown in blue. The Cα distance between the heavy-chain C-terminal residues of the symmetry-related Fabs is 58 Å and is marked by dashed lines. (F) Road map showing the density of AM28 Fab (red line contour; radius, 155 to 156 Å) and the epitopes HEWTPSWA (VP0; yellow), HQDKP (VP0; cyan), PLSIPTGSANQVD (VP0; magenta), MADSTTPSENHG (VP3; blue), ATTAPQSIVH (VP3; green), and FFPNATTDST (VP3; red). An asymmetric unit is marked by black lines.