Abstract
Understanding distinct gene expression patterns of normal adult and developing fetal human pancreatic α- and β-cells is crucial for developing stem cell therapies, islet regeneration strategies, and therapies designed to increase β-cell function in patients with diabetes (type 1 or 2). Toward that end, we have developed methods to highly purify α-, β-, and δ-cells from human fetal and adult pancreata by intracellular staining for the cell-specific hormone content, sorting the subpopulations by flow cytometry, and, using next-generation RNA sequencing, we report the detailed transcriptomes of fetal and adult α- and β-cells. We observed that human islet composition was not influenced by age, sex, or BMI, and transcripts for inflammatory gene products were noted in fetal β-cells. In addition, within highly purified adult glucagon-expressing α-cells, we observed surprisingly high insulin mRNA expression, but not insulin protein expression. This transcriptome analysis from highly purified islet α- and β-cell subsets from fetal and adult pancreata offers clear implications for strategies that seek to increase insulin expression in type 1 and type 2 diabetes.
Introduction
Diabetes is a critical medical issue, with an estimated 350 million individuals afflicted, a doubling in incidence over the last generation (1); it is a leading cause of adult blindness, kidney failure, amputation, lost work, and premature mortality (2,3). While diabetes is easily diagnosed by simple blood glucose measurements, it ultimately results from a loss of functional β-cell mass. We need to better understand the molecular mediators driving that loss and limited β-cell regeneration capacity (4,5). This knowledge gap exists because it has not previously been possible to study homogeneous, highly enriched endocrine cell populations from human islets. Recent studies have reported expression profiles on whole islets (6) and individual cell types using techniques like laser capture microdissection (7), or FACS-enriched β-cells identified by their zinc content (8) or cell surface epitopes (9). Each of these approaches has inherent limitations, including extensive processing, RNA degradation, and lack of strict cell type specificity. We initially developed a FACS technique (10) to isolate murine pancreatic islet cells based upon their cell-defining hormone content. Our improved technique overcomes the RNA compromise encountered by cell permeabilization and fixation. Transcriptomes for each isolated cell can now be assessed with high fidelity and sensitivity, as recently described for pancreatic progenitors and β-cells (11,12). We report the transcriptomes of highly purified human adult and fetal islet α- and β-cells. We find that α-cells express a large amount of insulin (INS) mRNA despite lacking any detectable INS protein, suggesting that α-cells may, under certain circumstances, possess the capacity to differentiate into INS-producing β-cells (13–15) with transformative therapeutic implications.
Research Design and Methods
Dissociation, Fixation, and Staining
Adult human islets were obtained from the Integrated Islet Distribution Program (IIDP) or Prodo Laboratories, Inc. The islet donors used in RNA sequencing (RNASeq) analysis (4–60 years of age) were of both sexes (five males, one female, one undefined), and of variable BMI values (21.5–37 kg/m2; Table 1), and diabetes had not been diagnosed in any of them. This study was deemed exempt from review (by the Institutional Review Board), as all samples were de-identified from deceased donors. Islet dissociation and intracellular antibody staining used a published protocol (10–12) with modifications, which included using TrypLE (Invitrogen) for dissociation and incubating antibodies with RNasin for 30 min prior to adult tissue staining. Anti-INS (Gallus Immunotech), anti-chicken allophycocyanin (Jackson ImmunoResearch) and Zenon (Invitrogen)-conjugated anti-glucagon (GCG) (Sigma-Aldrich) with Pacific Blue, and anti-somatostatin (SST) (LSBio) with Zenon Alexa Fluor 488 were used to stain β-, α-, and δ-cells, respectively.
Table 1.
Fetal and adult donor demographic information, β-cell/α-cell ratio, and RNA integrity
| Sample ID | Age | Sex | BMI | β-Cell/α-cell ratio | RIN |
Heat map ID no. | |
|---|---|---|---|---|---|---|---|
| α-Cells | β-Cells | ||||||
| Fetal | |||||||
| SE 4546 | 18.3 weeks | Unk | NA | 8.1 | 1 | ||
| SE 4574 | 18 weeks | Unk | NA | 1.44 | 7.9 | 2 | |
| SE 3029 | 19 weeks | Unk | NA | 7.7 | 3 | ||
| SE 3646, 4810 | 13, 13.3 weeks | Unk | NA | 2.94 | 8.1 | 4 | |
| SE 3650, 3648, 3647 | 13, 14, 14 weeks | Unk | NA | 6.9 | 5 | ||
| SE 4539 | 18 weeks | Unk | NA | 8.0 | 1 | ||
| SE 4574 | 18 weeks | Unk | NA | 7.4 | 2 | ||
| SE 3004 | 18 weeks | Unk | NA | 7.1 | 3 | ||
| SE 3054, 3634, 3635 | 12, 13.4, 14 weeks | Unk | NA | 6.4 | 4 | ||
| SE 3637, 3638 | 12, 14 weeks | Unk | NA | 6.0 | 5 | ||
| SE 3646, 4810 | 13, 13.3 weeks | Unk | NA | 6.2 | 6 | ||
| Adult | |||||||
| ZEP055 | 60 years | F | 26.5 | 1.7 | 5.6 | 5.3 | 1 |
| ZEQ024 | 19 years | M | 21.5 | 1.9 | NA | 3.6 | 2 |
| ZA2357 | 24 years | M | 29.2 | 1 | 4.1 | 4.9 | 3 |
| #4 | 4 years | Unk | Unk | 1.81 | 5.8 | 6.3 | 4 |
| AAI1303 | 53 years | M | 27.2 | 3.2 | 3.7 | 5.8 | 5 |
| AAIB402A | 30 years | M | 37 | 1.35 | 5.6 | 5.2 | 6 |
| AAHL164 | 18 years | M | 31 | 1.7 | 7.3 | 7.1 | 7 |
The age, sex, and BMI information for the adult donors and information on fetal donors from which α- and β-cells were purified is shown. The measured β-cell/α-cell ratios and the RIN values for the purified α- and β-cells are shown. The heat map identification number corresponds to the data in Supplementary Table 1. F, female; M, male; NA, not applicable; Unk, unknown.
Fetal tissues (12–18 gestational weeks) were obtained from Advanced Bioscience Resources or StemExpress (some tissues were pooled; Table 1). Pancreata were minced and incubated with dispase for 10 min at 37°C. The tissue was gently disrupted using a blunt 14-gauge needle and incubated for another 30–40 min with digestion stopped using EDTA (5 mmol/L). Fetal cells were filtered, washed twice with PBS containing 5 mmol/L EDTA, fixed, and covered for 10 min with a fixation and permeabilization solution (BD Biosciences) with 20 μL RNAseOUT (Invitrogen) and 10 μL RNA Later (Invitrogen) at 4°C. Fixed cells were washed twice with 1xBD wash buffer (BD Biosciences) with ultra–pure-grade BSA (0.2%, Invitrogen), and RNAseOUT. Antibodies against INS (Fitzgerald), GCG (Sigma-Aldrich), and SST (Genetex) were conjugated to Zenon Alexa Fluor 647, R-phycoerythrin, or Pacific Blue, respectively. Antibody cocktails (antibodies in 1xBD washing solution with 20 μL RNaseOUT) were added to cells and were incubated at 4°C in the dark with gentle shaking for 25–30 min. Cells were washed twice with 1xBD wash buffer with 0.2% BSA and RNaseOUT, and once in PBS, and were resuspended in PBS with RNaseOUT.
FACS
Adult and fetal cell preparations were sorted using a BD Biosciences FACS Aria II (in the Flow Cytometry Core Laboratory at the University of Massachusetts Medical School). Gating strategies are shown (Supplementary Fig. 1, adult islets, and Supplementary Fig. 2, fetal tissue). Cells were sorted into PBS containing 0.5% BSA and RNasin (Promega). For adult tissues, the purified α-cell+ and β-cell+ populations were resorted and quantified to determine the cell purity of each cell subset (>97% pure). RNA was extracted from enriched α-, β-, and δ-fetal cell types, and purity was assessed by quantitative RT-PCR (Supplementary Fig. 2I).
RNA Purification
Sorted adult cells were pelleted and digested using the Ambion RecoverAll Total Nucleic Acid Isolation Kit. RNA was quantified and analyzed for RNA Integrity Number (RIN) using the Agilent 2100 Bioanalyzer and 6000 Pico Chip. RIN values ranged from 3.6 to 7.1 (Table 1). There was no correlation between the RIN values and gene expression variability between samples (Supplementary Fig. 3).
Sorted fetal cells were pelleted and resuspended in TE buffer containing 0.5% SDS and 0.8 µL of 20 mg/mL Proteinase K (Roche). Cells were incubated at 65°C for 2 h, and total RNA was isolated using phenol/chloroform extraction, aqueous phase precipitation with isopropanol, and columns (Qiagen). RIN values ranged from 6.0 to 8.1 (Table 1).
Library Construction
For adult islet α- and β-cell subsets (samples 1–3 and without SST staining) and fetal β-cell populations, up to 100 ng RNA for each cell population was isolated. Libraries were constructed by RNA fragmentation, first- and second-strand cDNA synthesis, adaptor ligation, amplification, library validation, and ribosomal RNA removal by double-stranded nuclease digestion (Beijing Genomics Institute). Ninety-one base pair paired-end sequencing (adult) or 50 base pair single-end sequencing (fetal) was performed on the Illumina HiSeq 2000 and TruSeq platforms, respectively.
Total RNA isolated from fetal α-cell subsets (1 ng) and adult islet cell subsets (5 ng, samples 4–7) was processed using the NuGEN Ovation RNA-Seq System V2 and Ovation Ultralow DR Multiplex Systems 1–8. Quantitative and qualitative assessments of cDNA libraries were performed using an Agilent DNA 1000 chip. Libraries were sequenced using the Illumina HiSeq 2000 platform, and 20 million clean reads per sample were acquired.
RNASeq Analysis
Bowtie 2 (16) was used to remove reads aligning to rRNA sequences by allowing up to one mismatch with multiple mappers. Remaining reads were aligned by RSEM software (17) to a human transcript reference file generated using University of California, Santa Cruz, RefSeq annotations downloaded 10 May 2014. RSEM was then used to calculate total and normalized reads per gene using the following parameters, in addition to defaults: -n 2 for maximum number of mismatches; -e 70 for maximum sum of mismatch quality scores across the alignment; -l 25 for minimum read or insert length allowed; -chunkmbs 500 for memory allocated for best first alignment calculation; -p 4 for the number of threads to use; and -m 200 for alignment suppression if >200 valid alignments exist for a read.
Heat Map Analysis
To remove transcriptional noise created by minimally expressed genes, we analyzed only genes that had an average number of transcripts per million (TPM) of >25 in at least one cell type (adult or fetal: α or β), with 6,524 genes satisfying that condition. To control for different library construction methods, ComBat (18) removed the batch effect. The heat map dendrograms (columns) were created using hierarchical clustering; genes (rows) were clustered using the k-means algorithm (k = 9). All analyses were performed in R. Non-negative matrix factorization (NMF) was performed using the R NMF package (19) on three samples from each cell type (Supplementary Figs. 4 and 5 and Supplementary Tables 1–3).
Protein Immunofluorescence-RNA Fluorescence In Situ Hybridization Double Labeling
Dissociated islets (250,000 cells/well) were cultured at 37°C, in 5% CO2 on eight chamber slides (Falcon) in IIDP islet culture medium overnight. Cells were washed with PBS, fixed with 4% paraformaldehyde, permeabilized for 10 min with 0.5% Triton X-100/PBS, and incubated in 8% BSA/PBS for 1 h, and overnight at 4°C with antibodies to INS (Dako) and GCG (Santa Cruz Biotechnology) in 2% BSA/PBS. After washing with PBS/0.1% Tween-20, cells were incubated with secondary antibodies (anti-guinea pig Alexa Fluor 633 and anti-rabbit Alexa Fluor 546; Life Technologies) for 1 h at room temperature. Cells were then washed; postfixed with 4% paraformaldehyde; dehydrated by incubating with 70%, 90%, and 100% ethanol; and then air dried. Cells and hybridization solution (20 mmol/L Tris, pH 7.4, 60% formamide, and 0.1 µg/mL salmon sperm DNA) were warmed to 82°C for 5 min. A hybridization solution containing an Alexa Fluor 488–labeled peptide nucleic acid (PNA) probe (100 nmol/L) for human and rat INS (488-O-TCCACAATGCCACGCTTCT; PNABio) was added to the cells, which were coverslipped and incubated at 82°C for 5 min, then at 37°C for 90 min in a humidified chamber. Following hybridization, cells were washed twice for 10 min in ×2 saline sodium citrate buffer containing 0.1% Tween-20 at 55°C and once in ×2 saline sodium citrate at room temperature. Nuclei were stained with DAPI (300 nmol/L in PBS) for 10 min, washed twice in PBS, and mounted in Prolong Gold (Invitrogen). Fluorescent images were captured using a Leica TS SP2 confocal microscope. Potentially overlapping emissions were collected separately using sequential scanning.
Dot Blot of Whole Islets and Sorted α-, β-, and δ-Cells
Whole human islets or fixed FACS-sorted cells were lysed in PBS containing protease inhibitors (Roche) and 1% SDS. Aliquots containing 3,000 cells/sample were dotted onto a nitrocellulose membrane (Thermo Scientific) incubated with antibodies (rabbit anti-INS, rabbit anti-GCG, or mouse anti-SST [Santa Cruz Biotechnology]; rabbit anti-GAPDH [Cell Signaling Technology]; mouse anti-GAPDH [R&D Systems]; or rabbit anti-GLUT1 antibody [a gift from Dr. Anthony Carruthers, University of Massachusetts Medical School]), probed with horseradish peroxidase–conjugated secondary antibodies (Jackson ImmunoResearch), and visualized using the Fujifilm LAS-3000 Imager with SuperSignal Reagent (Pierce).
Results
Human Islet Cellular Subset Composition Is Highly Variable
Human adult islets were dissociated into single-cell suspensions, stained, and FACS sorted into distinct hormone-specific populations (Fig. 1A–D, gating strategy for INS, GCG, and SST protein expression, and Supplementary Fig. 1). For any individual’s islets, the GCG+ α-cell composition ranged from 23.8% to 71.6%, and INS+ β-cell composition ranged from 28.4% to 76.2%, with the average adult islet having 1.42 times as many β-cells per α-cell. Adult islet SST+ δ-cells comprised a smaller proportion (2.4–12% of labeled cells). Although adult islet composition varied among donors (Fig. 1E) (20), the cellular constituency was not significantly influenced by age (Fig. 2A) or sex (Fig. 2B). Although not significant, the β-cell-to-α-cell ratio appears to decline with increasing BMI (Fig. 2C).
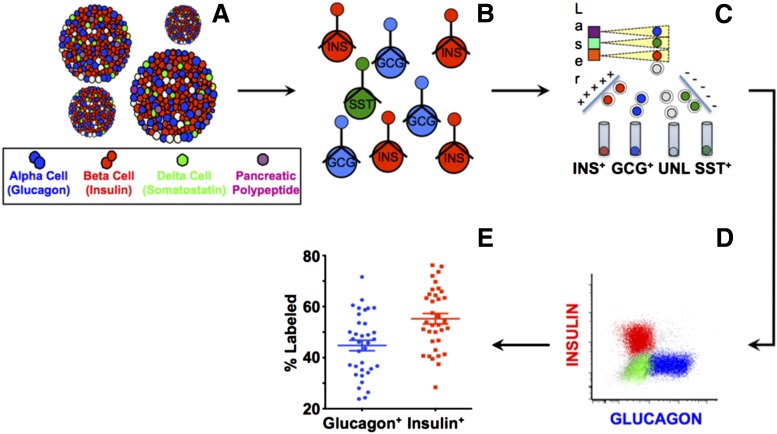
Figure 1.
Schema of islet cell subset separation and frequencies of donor islet cell subsets. A: Islets are composed of a heterogeneous mixture of α-cells (GCG+), β-cells (INS+), and δ-cells (SST+). Islets are dissociated into a single-cell suspension (B), fixed and permeabilized to stain for intracellular hormone content (C), and sorted using a FACS (D) into distinct populations. E: Composition of endocrine islet populations of α- and β-cells differs from donor to donor.
Figure 2.
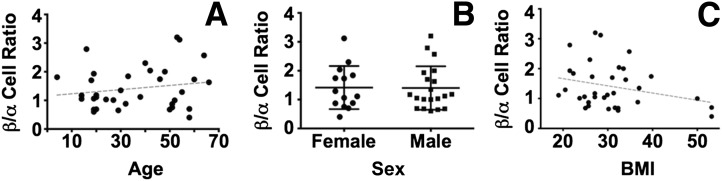
β-Cell/α-cell ratios do not differ with age, sex, or BMI. The percentage of α-, β-, and δ-cells were calculated for 34 human islet samples, as in Fig. 1. The relative ratios of β-cells to α-cells were calculated for each donor (average 1.42) and plotted against age (A), sex (B), and BMI (C). Linear regression analysis was performed for plots A and C (dashed line). No difference in β-cell/α-cell ratio can be attributed to age (Pearson r = 0.17 [95% CI −0.17 to 0.48]) or sex (unpaired P value = 0.95 [95% CI for SEM −0.55 to 0.52]). There is a trend toward a negative correlation of β-cell/α-cell ratio with increased BMI, but the values are not significant (Pearson r −0.27 [95% CI −0.56 to 0.07]).
Fetal pancreata do not contain defined islets; therefore, dissociated whole tissue was stained for endocrine hormones (Supplementary Fig. 2): INS+ β-cells accounted for ∼50%, GCG+ α-cells accounted for 20%, and SST+ δ-cells accounted for 30% of all sorted endocrine cells from fetal pancreata. The average ratio of INS+ cells per GCG+ cell was 2.5 (range 1.9–4.3), and per SST+ cell, it was 1.7 (range 1.5–1.9).
α- and β-Cell Transcriptome Signatures
From 10,000 adult islet equivalents (IEQs), we routinely isolated 200,000 β-cells and 150,000 α-cells, which provided ∼200 and 150 ng RNA, respectively. Assuming 500–1,000 cells/IEQ, that ∼50% of the cells are labeled, and that β-cells comprise (on average) ∼50% of the labeled cells (thus, the total β-cell number in 10,000 IEQs would be ∼2.5e6), our β-cell yield of ∼2.0e5 represents a cell recovery of ∼8% of the starting β-cells in the isolated islets. Fetal hormone+ cell yields depended on donor age. We obtained, on average, 6,811 INS+ cells (range 1,065–16,683 cells) from tissue up to 14 weeks' gestation (n = 11), and 44,530 (range 11,866–112,153 cells) from tissues of 18–21 weeks' gestation (n = 15). We isolated sufficient RNA for library construction and next-generation sequencing for seven adult β-cell and six adult α-cell samples (one α-cell yield was insufficient for analysis), and for five fetal α-cell and six fetal β-cell samples. All reads were processed using RSEM (21) with the RefSeq annotations (17). We deposited RNA sequence data in the GEO database (http://www.ncbi.nlm.nih.gov/geo/).
After normalizing to control for different library construction methods, hierarchical clustering of samples recapitulated sample origin. We used k-means clustering to find the characteristic gene expression patterns of each cell group (Fig. 3). Each of the clusters defined gene sets with high relative expression in specific cell types compared with other populations. In addition to genes that are specific to β- and α-cells, we found genes with distinct expression in adult versus fetal cells, and genes specific to developing fetal islet cells. Two clusters did not have a clear pattern of expression. RNA expression, heat map cluster, and TPM data are shown (Supplementary Tables 1–3 and Supplementary Figs. 4 and 5).
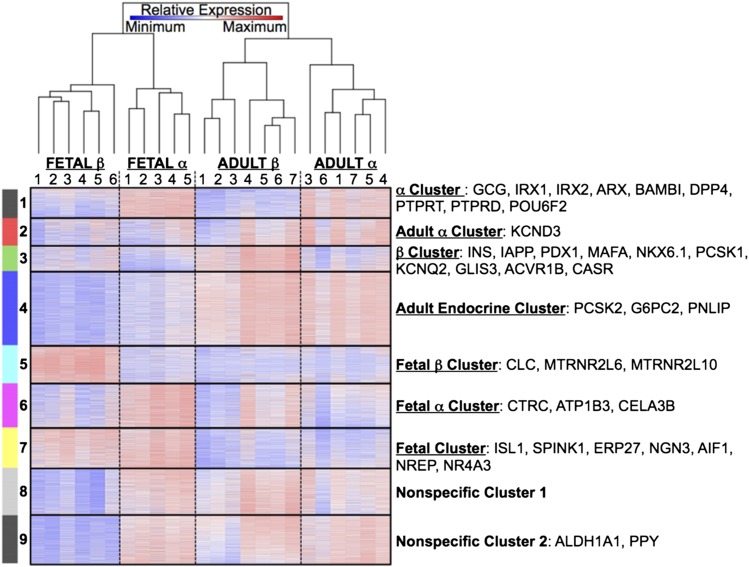
Figure 3.
The α- and β-cell gene expression patterns: heat map analysis. The cluster and heat map diagram shows the relative gene expression (blue represents minimal expression; white shows neutral expression; red indicates high gene expression) for sorted fetal human β-cells (n = 6; INS+), adult human β-cells (n = 7; INS+), adult human α-cells (n = 6; GCG+), and fetal human α-cells (n = 5; GCG+) separated by population. The sample dendrogram (columns) was calculated using hierarchical clustering, while the genes (rows) were clustered using the k-means algorithm. The specific genes listed to the right of each cluster demonstrate high relative gene expression in the cluster-specific samples compared with expression levels in all other samples.
Established and Enriched Transcripts in Highly Purified α- and β-Cell Populations
The unbiased RNASeq approach showed mRNA levels for genes commonly associated with each endocrine cell population, and was consistent with previous work that compared the expression of β- and non–β-cells (8) or β- and α-cells (13). For example, we found GCG and ARX (Aristaless related homeobox, an X-linked transcription factor) to be disproportionately expressed in α-cells (Fig. 3 and Supplementary Tables 1–3). This approach identified additional genes significantly disproportionately expressed in α-cells for which detailed functional relevance is not known: IRX1 and IRX2 (Iroquois homeobox transcription factors 1 and 2), DPP4 (dipeptidyl peptidase IV), PTPRT/PTPRD (protein tyrosine phosphatase receptor type T and D), POU6F2 (POU domain class 6, transcription factor 2), KCND3 (voltage-gated potassium channel Kv4.3), and BAMBI (activin membrane-bound inhibitor).
Similarly, several established β-cell genes were robustly and disproportionately expressed in the β-cell–specific cluster (Fig. 3 and Supplementary Tables 1–3): INS, IAPP (insulinoma amyloid polypeptide), PCSK1 (prohormone convertase 1), and transcription factors PDX1 (pancreatic and duodenal homeobox 1), MAFA (V-maf musculoaponeurotic fibrosarcoma oncogene homolog A), and NKX6.1 (NK6 homeobox 1) (22). Additionally, β-cells disproportionately expressed mRNA for genes of unknown relevance: GLIS3 (GLIS family zinc finger 3), ACVR1B (activin receptor like kinase 4), CASR (calcium sensing receptor), and KCNQ2 (potassium channel voltage-gated subfamily Q member 2).
Expression of Genes in Adult Versus Fetal Highly Purified α- and β-Cell Populations
Several genes implicated in early development are detectable in fetal but not adult islet cells (Fig. 3 and Supplementary Tables 1–3). Our assays detected high fetal, but not adult, β-cell expression of ISL1 (ISL Homeobox 1) and NGN3 (Neurogenin 3). Surprisingly, we found that developmental markers identified as fundamental for endocrine cell specification in the mouse (PRDM16 [PR Domain Containing 16] and PTF1A [Pancreas-Specific Transcription Factor 1]) were undetectable in human islet cells from donors who were 12–14 weeks old. Although islet cell subset expression profiles are highly variable across individuals (Fig. 3), genes associated with immune function, and in particular inflammatory genes, were highly enriched in fetal β-cells (P < 1.3 × 10−13 with a Benjamini-Hochberg correction). Examples of mRNA highly and disproportionately expressed in most fetal β-cell samples include the following: PTGS2 (Prostaglandin-Endoperoxide Synthase 2), a classic inflammatory enzyme; S100A9 (S100 Calcium Binding Protein), which has been associated with cystic fibrosis and been shown to promote leukocyte recruitment (23); and DEF1, DEFA1B, and DEFA4 (Defensin α 1, Defensin α 1B, and Defensin α 4) (Fig. 3 and Supplementary Tables 1–3).
Genome-Wide Association Studies Diabetes Genes
Genome-wide association study screens have identified >80 total genes associated with type 1 diabetes (Supplementary Fig. 6), type 2 diabetes (Supplementary Fig. 7), or INS and GCG expression (Supplementary Fig. 8) (24–28). Gene expression levels for each detected gene within the four sample populations show that >50% of these genes are expressed in either adult or fetal α- and β-cells, with an overabundance of DLK1 (Δ Like Homolog 1), GLIS3, and MEG3 (Maternally Expressed 3) found in β-cell populations. From a list of genes upregulated in islets from patients with type 2 diabetes (29), we found, with the exception of RPL36 (ribosomal protein L36) and COX5A (cytochrome c oxidase, subunit Va), that the reported genes were minimally expressed in purified α- and β-cells from donors in whom type 2 diabetes had not been diagnosed.
Hormone “Positive Selection” Increases Cell Type Purity
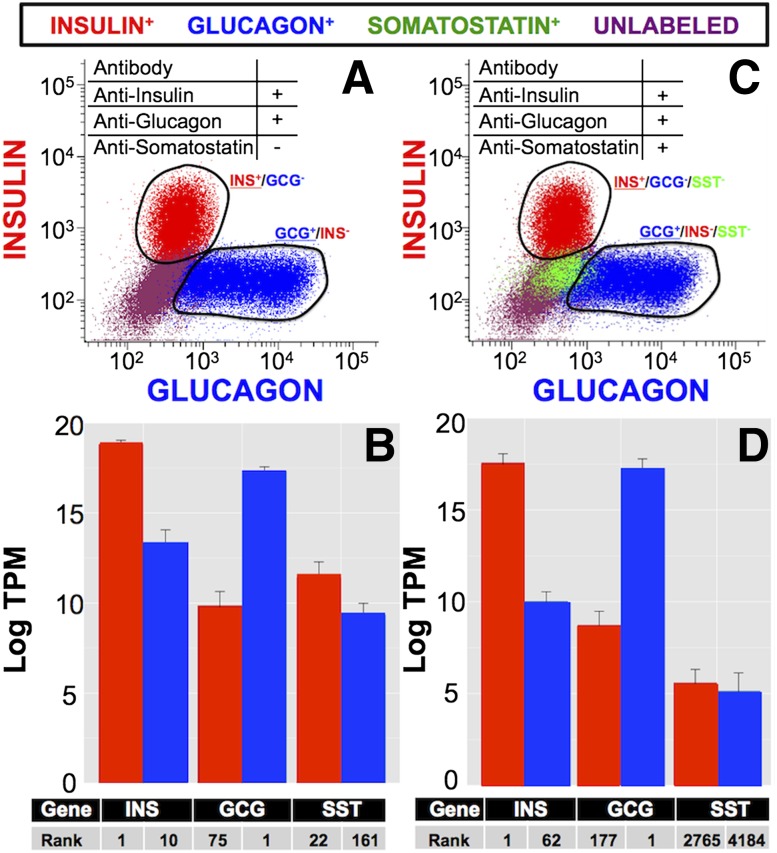
We compared the RNA sequencing results using two cell-sorting methods to assess the importance of cell purity for transcriptome analysis. In one method, we stained and sorted cells for all three hormones, sequenced the purified populations, and then compared these results to cells stained and sorted for INS and GCG, but not for SST. When SST+ cells were not removed from the sorted populations (Fig. 4A), SST mRNA gene expression appeared to be much higher in putative α- and β-cells (gene ranks of 161 and 22, respectively; Fig. 4B). When islet cells were stained for all three hormones (Fig. 4C), SST mRNA expression in the putative α- and β-cell populations was markedly decreased (gene ranks 2,765 and 4,184, respectively; Fig. 4D). Although SST+ cells comprise only >1% of the β-cell populations (data not shown), this contamination increased gene rank by >2,500 positions, emphasizing the importance of techniques that maximize cell type specificity for transcriptome analysis.
Figure 4.
Positive isolation of islet cell populations from adult islets increases cell type purity. Dissociated islets were analyzed as in Supplementary Fig. 1. Scatter plots for each population are shown (INS+ cells, red; GCG+ cells, blue; SST+ cells, green; unlabeled cells, purple). Transcriptome sequencing and gene expression were computed in TPM as described. Hormone staining for INS and GCG only (A) showed increased amounts of other hormones in the purified populations (B). Triple hormone staining that removes SST+ cells from the INS+ and GCG+ populations (C) provides a more specific overall gene expression profile for each cell type (D). The gene rank for each hormone is number 1 in each respective cell type (INS number 1 in INS+ cells; GCG number 1 in GCG+ cells). When SST protein is gated out of the sorted cells, the gene rank drops from 22 (B) to 2,765 (D) in INS+ cells (red), and from 161 (B) to 4,184 (D) in GCG+ cells (blue).
We built four expression profiles using NMF (30), corresponding to four cell populations (adult and fetal α- and β-cells). These profiles can be used to assess cell subset purity, as every sample is described as a linear combination of the four profiles. These results also highlight the significance of positive hormone selection. When SST is stained and gated out, subsequent α- and β-cell populations are more homogenous, which is evidenced by the higher coefficient of the adult α-cell and adult β-cell profiles (Supplementary Fig. 4). Staining for each hormone prevents gene expression contributions from contaminating cell types.
Many Human α-Cells Express Abundant INS mRNA, but No Detectable INS Protein
INS mRNA is the most abundantly expressed gene in human β-cells. Surprisingly, INS mRNA was also abundant in human α-cells, as the 62nd most highly expressed mRNA. This varies from GCG mRNA expression, which is number 1 in α-cells, but is number 177 in β-cells. With cell populations >97% pure, we sought to confirm the INS mRNA signal in α-cells using fluorescence in situ hybridization and protein quantitation to directly probe localized INS mRNA and protein expression in individual cells.
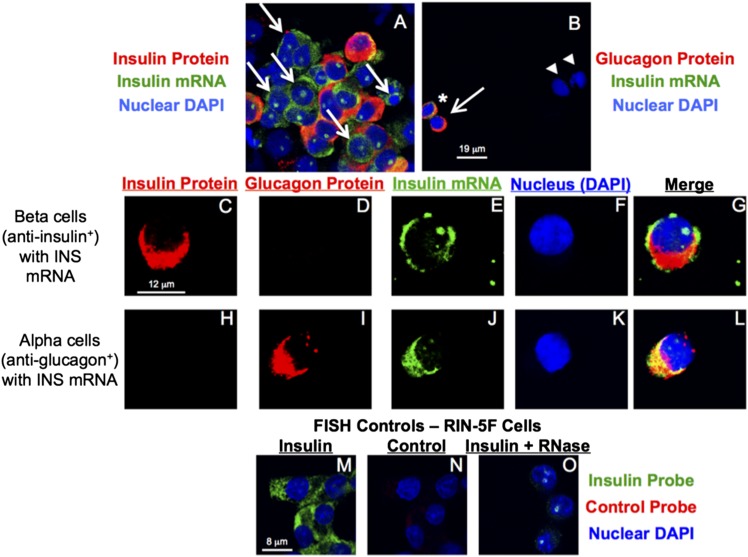
Dissociated islets were cultured on a slide and probed for INS and GCG protein and INS mRNA. Islet clusters contain distinctly positive signals for INS and GCG protein and INS mRNA (Fig. 5A and B). In β-cells (Fig. 5C–G), INS protein and mRNA are robustly expressed, while GCG protein is not detected. α-Cells (Fig. 5H–L) express GCG protein, as expected; but, in a substantial subset of those cells (39%), INS mRNA is also robustly expressed (Fig. 5L), confirming the α-cell INS mRNA expression rank determined by RNASeq. Control samples from an INSoma cell line show cells with specificity for the INS PNA probe (Fig. 5M–O).
Figure 5.
α-Cells express INS mRNA. Dissociated islets were stained with an antibody to INS and with a PNA probe to INS mRNA. Two islet clusters are shown. A: Arrows indicate INS mRNA+ cells that are negative for INS protein staining. B: As an internal control, islet cells show INS mRNA and GCG protein (*), GCG protein without INS mRNA (arrow), and two cells without GCG protein or INS mRNA message (arrowheads). Dissociated islets were stained with antibodies to INS (C–G) and GCG (H–L), and with a PNA probe to INS mRNA. A representative β-cell is shown that is positive for INS protein and message, but negative for GCG protein (C and D). A representative GCG+ α-cell is shown that is negative for INS protein (H), but positive for INS mRNA (I). Control RIN-5F cells all stain positive for INS mRNA (M), negative for the control probe (N), and with markedly diminished intensity following RNase treatment (O).
To further test the apparent α-cell expression of INS mRNA without corresponding INS protein, we created cell lysates from whole islets and each hormone-specific, FACS-sorted population, then analyzed protein expression via dot blot using different antibodies than used in the FACS (Fig. 6). Whole-islet INS and GCG protein expression is high, with much lower SST protein expression (consistent with islet composition). Each purified population displayed enriched staining for its specific hormone. The unlabeled population, from which INS+, GCG+, or SST+ cells were removed, did not express detectable levels of the hormone proteins.
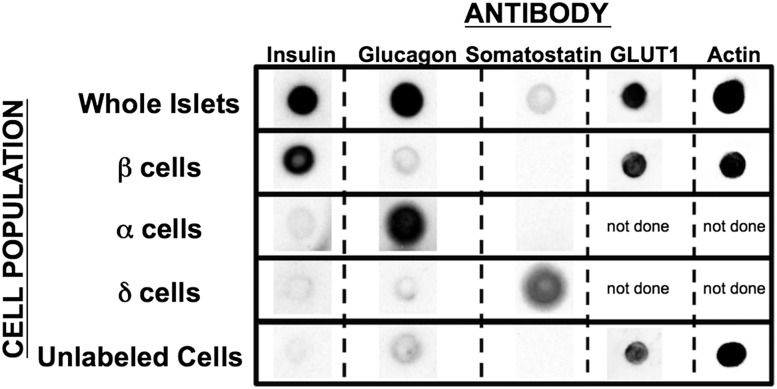
Figure 6.
Hormone protein expression in purified islet cell populations. A total of 3,000 cells were used for protein expression (INS, GCG, SST, β-actin, and GLUT1) determination via dot blot (representative image of three different replicates) as described. Staining was examined in whole islets; the three purified α-, β-, and δ-cell populations; and unlabeled cells, which did not stain for any of the three hormones. Each hormone is detected in the whole islet preparation, and its intensity increased within each respective purified cell population (INS in β-cells, GCG in α-cells, and SST in δ-cells).
Discussion
This report describes transcriptome profiles of α- and β-cells purified from human adult and fetal pancreata. We characterize pancreatic islet cellular constituents using a FACS, and compare human fetal and adult α- and β-cell subtype-specific gene expression profiles. Furthermore, we report that highly enriched human GCG+ α-cells express high levels of INS mRNA, but no detectable INS protein.
Brissova et al. (31) used confocal microscopy to observe the widely variable human islet α- and β-cellular proportions in nondiabetic donors. We confirm that variability using the FACS. These results support the use of human islets that were collected after cold ischemia and were cultured prior to shipping, but those same factors likely reduce the yield of α- and β-cells after dissociation and sorting, which may contribute to the RNASeq variability from donor to donor. We have expanded upon previous sorting and sequencing reports that used whole islets (6), Newport Green staining (8), cell surface selection (13), and hormone staining focusing only on β-cells (11,12), with methods to purify fetal and adult cells that contain INS, GCG, or SST, and by replacing nuclease protection assays (10) with RNASeq. Although fixation and permeabilization preclude downstream functional biochemical studies, we have used these improved techniques to examine gene expression in adult human α- and β-cells, and human fetal α- and β-cell populations, which will provide future targets for functional studies.
The unbiased techniques identified genes that are commonly associated with α- and β-cells, confirming our isolation methods. In addition to expected α-cell GCG mRNA expression, we confirmed the markedly higher ARX expression, previously reported from mice (32,33) and humans (34) to maintain α-cell fate. More interestingly, we observed robust fetal and adult human α-cell IRX1 and IRX2 expression, previously implicated only through studies (35) of NGN3 mutant embryonic mice with abnormal α-cell development. From those mouse studies, the IRX1 and IRX2 transcription factors were observed in the wild-type, but not the mutant, mouse endoderm. Human β-cell GLIS3 mRNA expression is noted because rodent studies have found that GLIS3 appears to play an important role in β-cell development and function, and it has been associated with type 1 diabetes in humans by genome-wide association studies (8,24,25,28,36,37).
We observed highly expressed mRNAs (ACVR1B) in purified adult islet cell populations for which detailed function is not described. ACVR1B is a single-transmembrane domain serine/threonine kinase receptor that binds activin. Nodal ligands (like activin) signal through ACVR1B and are thought to act as mesendoderm inducers during the generation of embryonic stem cells into pancreatic β-cells (38).
Tissue availability has limited early mouse pancreatic embryogenesis RNASeq studies, which precede β-cell expansion. Human fetal (12- to 18-week-old) pancreatic α- and β-cell transcriptional profiling offers insight into early islet development. If at 12–14 weeks' gestation, fetal pancreas mRNA (ISL1 and NGN3) levels mirror transcription factor levels, our data suggest that the expression of certain developmental factors may be transient and necessary only to trigger, but not to maintain, islet cell identity. We hypothesize that α- and β-cell progenitors commit very early in development, and maintain that cell lineage commitment into adulthood. Factors underlying the early progenitor commitment could provide a partial explanation for the widely variable β-cell mass that is now widely reported (31). We also report the fetal β-cell expression of inflammatory gene mRNAs DEFA1, DEFB1, DEFA4, PTGS2, and S100A9; previous studies (12) have reported robust human α-cell defensin mRNA expression.
Previous studies (39) using mouse islets showed that endocrine cells expressed multiple hormones in a single cell. We extended this observation by noting high INS mRNA expression in GCG protein expressing human α-cells. We did not find similarly high GCG mRNA expression in human β-cell subsets. The α-cell INS mRNA expression that we observe matches published results using other techniques (8,13), but those authors concluded that the apparent α-cell INS mRNA expression was secondary to β-cells contaminating their α-cells. We support these observations using fluorescence in situ hybridization, which found that approximately one-third of α-cells express INS mRNA, likely representing a unique α-cell subset. Our results are consistent with previous observations (40) that bulk sequencing masks single-cell transcript expression heterogeneity in purified T-lymphocyte populations.
In summary, we report detailed transcriptomes of highly purified human fetal and adult α- and β-cells using RNASeq. We have observed that islet cellular composition (as measured by the β-cell/α-cell ratio) is not significantly influenced by age or sex, though a trend may exist for donors with higher BMI values displaying a decreased β-cell/α-cell ratio. We also report that many transcription factors implicated in β-cell development from rodent studies (ISL1, PAX4, and NGN3) are highly expressed in human fetal β-cells, but much less so in adult human β-cells. Interestingly, while the function of these gene products is unclear, human fetal β-cells (compared with fetal α-cells from the same donor or with adult endocrine cell subsets) highly express mRNAs that are traditionally associated with inflammatory gene products. Last, our study of very pure cell subsets has uncovered surprisingly high INS mRNA expression (without detectable INS protein) in GCG-expressing α-cells, which raises the intriguing possibility that, under certain conditions (13–15), α-cells may be reprogrammed to secrete INS with clear therapeutic consequences. Furthermore, FACS data show that α- and β-cells from the same donor display a 100-fold range of GCG and INS levels, showing that there are likely different cell subsets within each population. We emphasize the importance of reporting quantitative data with regard to isolated β-cell yields since it is possible that different purification methods may enrich different β-cell subsets. We are now applying these techniques to assess islet subcellular composition and gene expression from donors with type 1 and type 2 diabetes.
Supplementary Material
Article Information
Funding. This research was supported in part by National Institutes of Health grants U01-DK-089572 and UC4-DK-104218. D.M.H. was supported by the William and Doris Krupp Chair in Medicine.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. D.M.B. was responsible for the experimental design, carried out the experiments, was responsible for data analysis, and wrote the manuscript. A.N. was responsible for the experimental design, carried out the experiments, and wrote the manuscript. S.A. was responsible for data analysis and wrote the manuscript. S.P. and A.J.C. carried out the experiments. N.J.K., S.K., and P.d. were responsible for the experimental design and carried out the experiments. A.K. was responsible for data analysis. R.J.D. and D.L.G. were responsible for the experimental design. S.C.K. wrote the manuscript. M.G.G. and D.M.H. were responsible for the experimental design and wrote the manuscript. D.M.H. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Prior Presentation. Parts of this study were presented in abstract form at the 74th Scientific Sessions of the American Diabetes Association, San Francisco, CA, 13–17 June 2014; and the 2014 Rachmiel Levine Diabetes and Obesity Symposium: Advances in Diabetes Research, Pasadena, CA, 12–15 March 2014.
RNA sequence data will be deposited in the GEO database (http://www.ncbi.nlm.nih.gov/geo/).
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db15-0039/-/DC1.
D.M.B., A.N., and S.A. contributed equally to this work.
M.G.G., D.M.H., and P.d. are co-senior authors.
Deceased.
References
- 1.Danaei G, Finucane MM, Lu Y, et al.; Global Burden of Metabolic Risk Factors of Chronic Diseases Collaborating Group (Blood Glucose) . National, regional, and global trends in fasting plasma glucose and diabetes prevalence since 1980: systematic analysis of health examination surveys and epidemiological studies with 370 country-years and 2·7 million participants. Lancet 2011;378:31–40 [DOI] [PubMed] [Google Scholar]
- 2.Mathers CD, Loncar D. Projections of global mortality and burden of disease from 2002 to 2030. PLoS Med 2006;3:e442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roglic G, Unwin N, Bennett PH, et al. The burden of mortality attributable to diabetes: realistic estimates for the year 2000. Diabetes Care 2005;28:2130–2135 [DOI] [PubMed] [Google Scholar]
- 4.van Belle TL, Coppieters KT, von Herrath MG. Type 1 diabetes: etiology, immunology, and therapeutic strategies. Physiol Rev 2011;91:79–118 [DOI] [PubMed] [Google Scholar]
- 5.Weir GC, Bonner-Weir S. Islet β cell mass in diabetes and how it relates to function, birth, and death. Ann N Y Acad Sci 2013;1281:92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cnop M, Abdulkarim B, Bottu G, et al. RNA sequencing identifies dysregulation of the human pancreatic islet transcriptome by the saturated fatty acid palmitate. Diabetes 2014;63:1978–1993 [DOI] [PubMed] [Google Scholar]
- 7.Marselli L, Thorne J, Ahn YB, et al. Gene expression of purified beta-cell tissue obtained from human pancreas with laser capture microdissection. J Clin Endocrinol Metab 2008;93:1046–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nica AC, Ongen H, Irminger JC, et al. Cell-type, allelic, and genetic signatures in the human pancreatic beta cell transcriptome. Genome Res 2013;23:1554–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorrell C, Abraham SL, Lanxon-Cookson KM, Canaday PS, Streeter PR, Grompe M. Isolation of major pancreatic cell types and long-term culture-initiating cells using novel human surface markers. Stem Cell Res (Amst) 2008;1:183–194 [DOI] [PubMed] [Google Scholar]
- 10.Pechhold S, Stouffer M, Walker G, et al. Transcriptional analysis of intracytoplasmically stained, FACS-purified cells by high-throughput, quantitative nuclease protection. Nat Biotechnol 2009;27:1038–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hrvatin S, Deng F, O’Donnell CW, Gifford DK, Melton DA. MARIS: method for analyzing RNA following intracellular sorting. PLoS ONE 2014;9:e89459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hrvatin S, O’Donnell CW, Deng F, et al. Differentiated human stem cells resemble fetal, not adult, β cells. Proc Natl Acad Sci U S A 2014;111:3038–3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bramswig NC, Everett LJ, Schug J, et al. Epigenomic plasticity enables human pancreatic α to β cell reprogramming. J Clin Invest 2013;123:1275–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mezza T, Muscogiuri G, Sorice GP, et al. Insulin resistance alters islet morphology in nondiabetic humans. Diabetes 2014;63:994–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang YP, Thorel F, Boyer DF, Herrera PL, Wright CV. Context-specific α-to-β-cell reprogramming by forced Pdx1 expression. Genes Dev 2011;25:1680–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012;9:357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011;12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012;28:882–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaujoux R, Seoighe C. A flexible R package for nonnegative matrix factorization. BMC Bioinformatics 2010;11:367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim A, Miller K, Jo J, Kilimnik G, Wojcik P, Hara M. Islet architecture: a comparative study. Islets 2009;1:129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pruitt KD, Brown GR, Hiatt SM, et al. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res 2014;42:D756–D763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cano DA, Soria B, Martín F, Rojas A. Transcriptional control of mammalian pancreas organogenesis. Cell Mol Life Sci 2014;71:2383–2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Croce K, Gao H, Wang Y, et al. Myeloid-related protein-8/14 is critical for the biological response to vascular injury. Circulation 2009;120:427–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barrett JC, Clayton DG, Concannon P, et al.; Type 1 Diabetes Genetics Consortium . Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet 2009;41:703–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morris AP, Voight BF, Teslovich TM, et al.; Wellcome Trust Case Control Consortium; Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) Investigators; Genetic Investigation of ANthropometric Traits (GIANT) Consortium; Asian Genetic Epidemiology Network–Type 2 Diabetes (AGEN-T2D) Consortium; South Asian Type 2 Diabetes (SAT2D) Consortium; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium . Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet 2012;44:981–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jain P, Vig S, Datta M, et al. Systems biology approach reveals genome to phenome correlation in type 2 diabetes. PLoS One 2013;8:e53522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bergholdt R, Brorsson C, Palleja A, et al. Identification of novel type 1 diabetes candidate genes by integrating genome-wide association data, protein-protein interactions, and human pancreatic islet gene expression. Diabetes 2012;61:954–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elbein SC, Gamazon ER, Das SK, Rasouli N, Kern PA, Cox NJ. Genetic risk factors for type 2 diabetes: a trans-regulatory genetic architecture? Am J Hum Genet 2012;91:466–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pullen TJ, Rutter GA. When less is more: the forbidden fruits of gene repression in the adult β-cell. Diabetes Obes Metab 2013;15:503–512 [DOI] [PubMed] [Google Scholar]
- 30.Brunet JP, Tamayo P, Golub TR, Mesirov JP. Metagenes and molecular pattern discovery using matrix factorization. Proc Natl Acad Sci U S A 2004;101:4164–4169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brissova M, Fowler MJ, Nicholson WE, et al. Assessment of human pancreatic islet architecture and composition by laser scanning confocal microscopy. J Histochem Cytochem 2005;53:1087–1097 [DOI] [PubMed] [Google Scholar]
- 32.Collombat P, Mansouri A, Hecksher-Sorensen J, et al. Opposing actions of Arx and Pax4 in endocrine pancreas development. Genes Dev 2003;17:2591–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dhawan S, Georgia S, Tschen SI, Fan G, Bhushan A. Pancreatic β cell identity is maintained by DNA methylation-mediated repression of Arx. Dev Cell 2011;20:419–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spijker HS, Ravelli RB, Mommaas-Kienhuis AM, et al. Conversion of mature human β-cells into glucagon-producing α-cells. Diabetes 2013;62:2471–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petri A, Ahnfelt-Rønne J, Frederiksen KS, et al. The effect of neurogenin3 deficiency on pancreatic gene expression in embryonic mice. J Mol Endocrinol 2006;37:301–316 [DOI] [PubMed] [Google Scholar]
- 36.Kang HS, Kim YS, ZeRuth G, et al. Transcription factor Glis3, a novel critical player in the regulation of pancreatic beta-cell development and insulin gene expression. Mol Cell Biol 2009;29:6366–6379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nogueira TC, Paula FM, Villate O, et al. GLIS3, a susceptibility gene for type 1 and type 2 diabetes, modulates pancreatic beta cell apoptosis via regulation of a splice variant of the BH3-only protein Bim. PLoS Genet 2013;9:e1003532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johannesson M, Ståhlberg A, Ameri J, Sand FW, Norrman K, Semb H. FGF4 and retinoic acid direct differentiation of hESCs into PDX1-expressing foregut endoderm in a time- and concentration-dependent manner. PLoS One 2009;4:e4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katsuta H, Akashi T, Katsuta R, et al. Single pancreatic beta cells co-express multiple islet hormone genes in mice. Diabetologia 2010;53:128–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Newell EW, Davis MM. Beyond model antigens: high-dimensional methods for the analysis of antigen-specific T cells. Nat Biotechnol 2014;32:149–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.