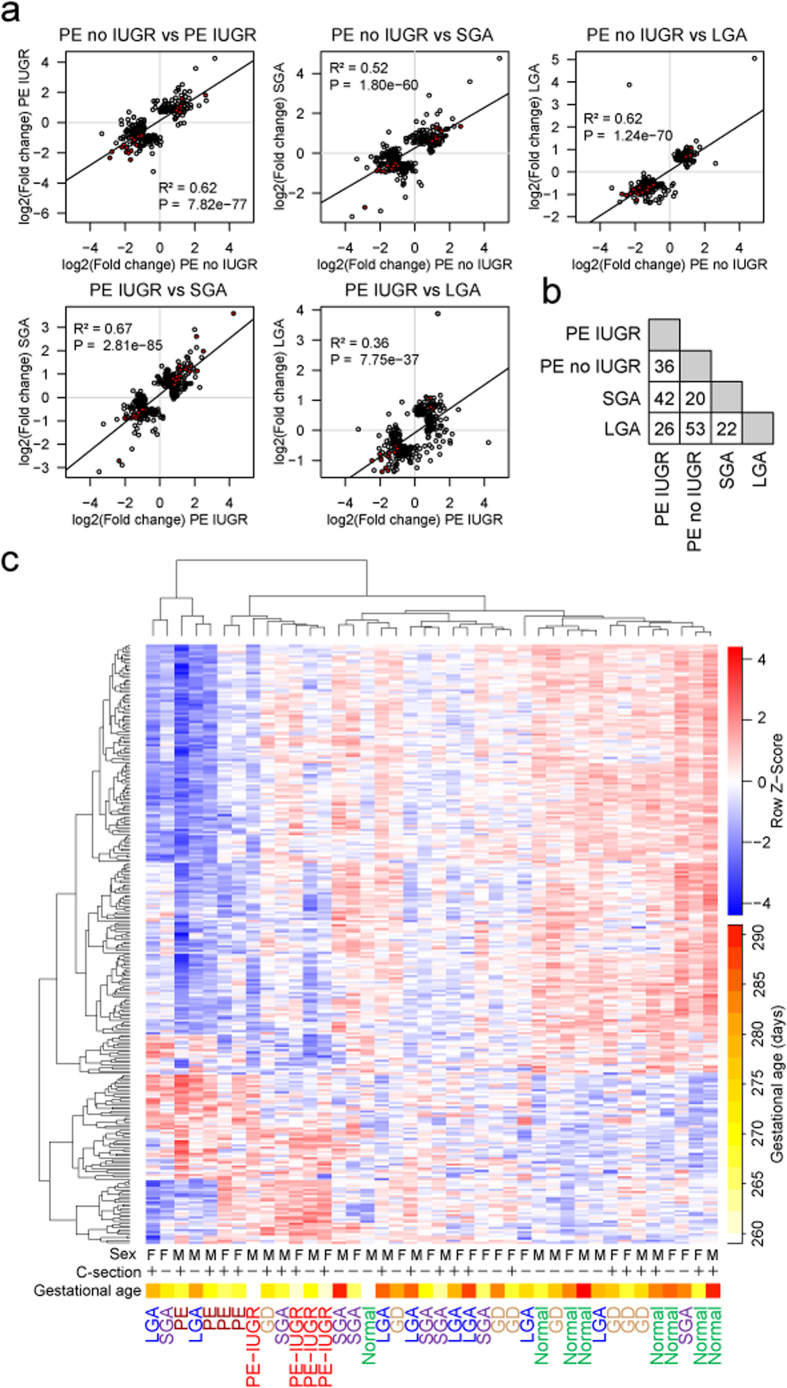
Figure 6. Placentas from the cases of late-onset preeclampsia (PE) with and without concomitant intra-uterine growth restriction (IUGR) exhibit distinct gene expression patterns.
(a) Correlation plots for the fold changes of the highest ranked genes in the differential expression testing in each pregnancy complication compared to normal pregnancy (DESeq analysis). For each pairwise analysis of gestational complications, the lists of top-200 genes (circles) were united and plotted at the x,y-plane, where the axes correspond to log2(fold changes) in the two groups. Red circles represent genes, which are shared between the top gene lists. The linear regression line along with correlation coefficient R2 and statistical significance is given. (b) Numbers of shared genes among the top 200 highest ranked transcripts in differential expression testing. Detailed information on the pairwise overlaps among the study groups for the shared top-genes with altered placental expression is provided in Supplementary Fig. S4. (c) Hierarchical clustering based on transformed read counts of 283 differentially expressed genes in PE without IUGR, PE with IUGR, SGA, LGA and GD. Gene expression levels were subjected to variance stabilizing transformation in DESeq and standardized by subtracting the mean expression across all samples from its value for a given sample and then dividing by the standard deviation across all the samples. This scaled expression value, denoted as the row Z-score, is plotted in red-blue color scale with red indicating increased expression and blue indicating decreased expression. Hierarchical clustering of genes (rows) and samples (columns) was based on Pearson’s correlation. Hierarchical clustering trees are shown for the analyzed samples (top) and genes (left). For each sample are shown newborn sex (M, male; F, female), delivery by caesarean section (+/–) and gestational age at birth plotted in white-yellow-red color scale (white < 260, red > 290 gestational days).