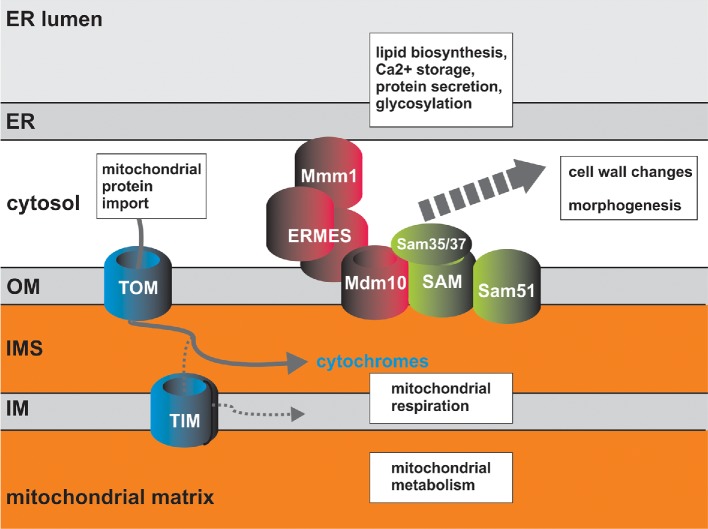
Figure 1.
The mitochondrial biogenesis apparatus in C. albicans. Proteins required for mitochondrial protein import are shown. All components of the mitochondrial import apparatus as described in the model baker's yeast can be found by bioinformatics searches in C. albicans. TOM is the ‘gateway’ to mitochondria through which all imported proteins pass. TIM enables translocation into the matrix and targeting to the inner membrane. In C. albicans, some cytochromes (b2 and c1) follow a distinct route to the intermembrane space to what is known in S. cerevisiae. The pathways required for their mitochondrial import in C. albicans remain to be discovered. Candida albicans contains an additional Omp85 family member, Sam51, which is part of the SAM complex, and data suggest that SAM and ERMES are bridged together via Mdm10 (Victoria Hewitt, A.T. and Trevor Lithgow, in preparation). The Sam37 and Sam35 subunits of the SAM complex and entire ERMES complex are not conserved in humans, and Sam37 and Mmm1 have been found to be required for virulence of C. albicans in the systemic mouse model (Becker et al. 2010; Qu et al. 2012). Links between SAM and ERMES function and cell wall integrity and hyphal morphogenesis have been made (Dagley et al. 2011; Qu et al. 2012), reenforcing the concept that mitochondria have broad impact on pathways required for fungal pathogenesis.