Abstract
Premise of the study:
Phylogenetic inference is moving to large multilocus data sets, yet there remains uncertainty in the choice of marker and sequencing method at low taxonomic levels. To address this gap, we present a method for enriching long loci spanning intron-exon boundaries in the genus Heuchera.
Methods:
Two hundred seventy-eight loci were designed using a splice-site prediction method combining transcriptomic and genomic data. Biotinylated probes were designed for enrichment of these loci. Reference-based assembly was performed using genomic references; additionally, chloroplast and mitochondrial genomes were used as references for off-target reads. The data were aligned and subjected to coalescent and concatenated phylogenetic analyses to demonstrate support for major relationships.
Results:
Complete or nearly complete (>99%) sequences were assembled from essentially all loci from all taxa. Aligned introns showed a fourfold increase in divergence as opposed to exons. Concatenated analysis gave decisive support to all nodes, and support was also high and relationships mostly similar in the coalescent analysis. Organellar phylogenies were also well-supported and conflicted with the nuclear signal.
Discussion:
Our approach shows promise for resolving a recent radiation. Enrichment for introns is highly successful with little or no sequencing dropout at low taxonomic levels despite higher substitution and indel frequencies, and should be exploited in studies of species complexes.
Keywords: COS marker, Heuchera, intron enrichment, Saxifragaceae, targeted enrichment
The necessity of multilocus data sets for phylogenetic analysis is now largely taken for granted. Low-copy nuclear sequences have long been advocated for the future of plant phylogenetic reconstruction, especially at low taxonomic levels where their high rate of evolution (particularly for introns) results in more-resolved phylogenetic hypotheses (Sang, 2002). These types of markers are also critical for accounting for phylogenetic discord across the genome, caused by such widespread phenomena as lineage sorting and hybridization. From the coalescent point of view, the canonical nonrecombining organellar genome (such as the typical angiosperm plastome, which has been heavily relied upon for developing Sanger markers) is essentially a single long phylogenetic marker with a single underlying history. This means that recent progress in the massive sequencing of entire organellar genomes (e.g., Ruhfel et al., 2014) does not necessarily equate in itself to adequate genomic sampling of phylogenetic signal for some questions, particularly at lower taxonomic levels, although such data sets may achieve high levels of phylogenetic resolution. The cost of whole genome shotgun sequencing has fallen dramatically in recent years, yet it remains costly to produce complete eukaryotic genomes at the scale typical for population and phylogenetic study (but see Cao et al., 2011), necessitating economical methods for broad subsampling of genomic loci. Genome skimming (Straub et al., 2012) is simple and can result in large amounts of rDNA, chloroplast, and mitochondrial data, yet this alone may not offer enough independent loci if either coalescent methods or the gene trees themselves are of interest.
Reduced-representation methods for next-generation sequencing (NGS) have emerged as a new standard for the collection of phylogenetic and population-genetic data. Restriction-based methodologies such as RADSeq are most commonly used for population studies, and targeted sequencing methods, usually based on target enrichment via synthesized RNA probes, have gained traction at higher levels for phylogenetics (McCormack et al., 2013, table 1; Cronn et al., 2012, table 5). For the taxonomic middle ground, at the level of species complex, appropriate methodologies remain unclear. While the best methodology may be lineage-specific to some extent, there remains a paucity of literature examining NGS methodology performance for these systems that would help inform the design of new sequencing projects. The application of restriction-based protocols at taxonomic levels beyond that of populations, or perhaps subspecific taxa, entails a greater risk due to the possibility that many restriction sites may possess mutations in a subset of the study samples, resulting in frequent allele dropout and, at worst, data matrices consisting largely of missing data. Homology assessment may also be more difficult if anonymous restriction-based markers are to be used for phylogenetics.
Targeted sequencing based on in-solution hybridization of DNA fragments to biotinylated RNA probes (Gnirke et al., 2009) avoids these problems. Moreover, hybridization-based methods can be combined with genome-skimming techniques to obtain nontargeted high-copy loci (i.e., “Hyb-Seq”; Weitemier et al., 2014), a benefit that generally is not shared by other techniques. On the other hand, probe-hybridization-based techniques have typically targeted exonic regions that may be too conserved among closely related species where Sanger markers have failed to obtain resolution, resulting in large amounts of uninformative data and therefore wasted sequencing effort, even if some degree of off-target intron assembly is possible (Weitemier et al., 2014). The performance of such a methodology in obtaining less conserved nonexonic sequences (such as introns or intergenic regions) is currently poorly known, but comparative genomic data in the form of closely related genome and transcriptome sequences are becoming increasingly available to facilitate the identification and use of such loci.
The genus Heuchera L. (Saxifragaceae), with about 43 species, appears to represent a recent, relatively rapid radiation for which these methodological decisions are especially critical. Deng et al. (2015) estimate the age of the “Heuchera group” (including related genera such as Tiarella L. and Mitella L.) to be 7.27 mya, a date recent enough to place the origin of many clades in Heuchera in the Pleistocene. Such recent divergence events are expected to leave few informative mutations for phylogenetic reconstruction, motivating the use of numerous and more quickly evolving regions. A recent study based on Sanger sequencing (Folk and Freudenstein, 2014) confirms the difficulty of resolving many of these relationships, particularly within taxonomic sections, reinforcing the need for careful marker choice.
We therefore aimed to evaluate the RNA-probe-based targeted sequencing of a panel of long, continuous loci containing both intron and exon sequences, using a combination of genomic and transcriptomic data. Because we were interested primarily in phylogenetic conflict, and therefore required resolved gene trees, we favored loci that were much longer than those typical in recent efforts. Using a Hyb-Seq approach (Weitemier et al., 2014), it is possible to obtain large amounts of organellar data from enrichment experiments, but the need for additional organellar genomes in the Saxifragaceae and other plant groups is acute given that there exists only one unpublished chloroplast genome for the family, and there is no mitochondrial genome available even for the Saxifragales. To develop further phylogenetic markers, we additionally sought to assemble organellar genomic resources for phylogenomics in the Saxifragaceae that should be of broad utility for related genera.
Aspects of target enrichment relating to wet-laboratory methods, off-target genome assembly, and phylogenetic reconstruction are now well covered in exon-based enrichment methods (Mandel et al., 2014; Weitemier et al., 2014). Some recent studies have also included intronic probes (e.g., Govindarajulu et al., 2015); our work contributes to this publication series by focusing on the effectiveness of intron enrichment and long-target enrichment for closely related species. While we were collecting these data, a study also including intron probes was published for Medicago L. (de Sousa et al., 2014); these authors also point out the use of introns for lower-level taxonomic questions. However, this study only reports experimental statistics on entire loci—discussion and data on the effectiveness of intron enrichment are absent. Additionally, their locus design methodology cannot be used on the majority of organisms that lack a high-quality annotated reference genome, underlining the need for broader methodologies and data exploration for use by the phylogenomic community.
METHODS
Materials
The DNAs we used were primarily of high quality and prepared from cultivated material or quickly dried field material. However, we also successfully sequenced a herbarium specimen of H. acutifolia Rose, collected in 1984; we have had further success with specimens collected as long ago as the 1940s (unpublished). The same H. acutifolia sample that was successful for this study previously proved impossible to sequence for low-copy markers using a Sanger protocol (Folk and Freudenstein, 2014; similar observations in Cronn et al., 2012). As an initial test of our method, we present results from 15 individuals selected as taxonomically representative of the major sections of the genus (Appendix 1).
Genomic reference development
As a draft genomic reference, we used 3.7 Gbp of short-read data (20,288,896 reads) from H. parviflora var. saurensis R. A. Folk. Paired-end sequencing was performed by The Ohio State University Molecular and Cellular Imaging Center in six multiplexed runs with samples from unrelated projects (five Illumina MiSeq runs with 300-bp reads; one Illumina HiSeq lane with 100-bp reads; all data uploaded to the Short Read Archive, accession SRP058301). This level of sequencing corresponds to approximately 7.5× expected average nuclear genomic coverage using 2C values from Godsoe et al. (2013). While over-sampling of high-copy loci will reduce the actual coverage of the single-copy portion of the genome, most nuclear contigs consistently showed 5–8× coverage. These data were assembled in Velvet (Zerbino and Birney, 2008) using a kmer length of 64, expected insert length of 700 bp, standard deviation of insert length 100 bp, and minimum contig length of 1000, to develop reference contigs for the low-copy portion of the genome. This resulted in 18,344 contigs between 1000 and 7360 bp; a longer kmer length results in longer contigs, but these were not necessary for marker development. A BLAST search against organellar references confirmed that organellar and rDNA sequences were not present in this batch of contigs.
Because we wished to explore the possibility of off-target assembly of high-copy loci, we used the skimming data from H. parviflora var. saurensis to assemble complete chloroplast and mitochondrial genomes (GenBank accessions KR478645 and KR559021, respectively). The chloroplast genome was assembled by mapping all reads (trimmed with Geneious default settings) using the native Geneious assembler (version 7, Biomatters; available from http://www.geneious.com/) with medium sensitivity, using the chloroplast genome of H. sanguinea Engelm. (M. Moore, unpublished). The resultant contig was manually screened for regions of mis-mapped reads (always due to large indels in intergenic spacers); at these points the consensus sequence was broken into subsequences. These subsequences were subjected to iterative assembly in Geneious (25 iterations), repeated until all consensus sequences of these assemblies overlapped (by de novo assembly in Geneious with default settings) and could be combined into a single circle. The read mapping was repeated on this resultant genome draft, and the complete lack of mis-mapping regions was confirmed.
The mitochondrial assembly method was largely similar to that of the chloroplast; however, a reasonably close reference mitochondrial genome was not available for the initial read-mapping. Geneious de novo assembly was performed with default settings on the entire data set, divided into three approximately equal parts to reduce RAM requirements to feasible levels. Mitochondrial contigs were distinguished by BLAST searches against the Vitis vinifera L. mitochondrial genome (Goremykin et al., 2008). This procedure was found to produce very long mitochondrial contigs compared to preliminary Velvet and SOAP (Luo et al., 2012) assemblies, as long as 122,611 bp. Comparing the assemblies revealed that the Geneious contigs contained all of the mitochondrial sequence assembled by the other programs, but in fewer pieces. Consensus sequences of these contigs (n = 39) were used in iterative assembly, circular reference construction, and assembly validation as above.
Organellar genome annotations were performed using Geneious, based on H. sanguinea (chloroplast) or V. vinifera (mitochondrion); synteny comparisons were performed using the Mauve plug-in (Darling, 2004) in Geneious.
Locus design
To obtain putatively single-copy loci in the absence of comparative genomic data within Saxifragaceae, Velvet contigs (n = 18,334) were searched against a panel of conserved single-copy sequences (hereafter “COS loci”) from the Arabidopsis genome (Kozik et al., unpublished; but see http://www.cgpdb.ucdavis.edu/COS_Arabidopsis/; also used in Mandel et al., 2014; n = 3714) using BLAST, yielding 3251 hits that were retained for RNA transcript queries. It is important to note that for these retained loci, we had two sources of assurance of their low-copy (likely single-copy) status: (a) they are single-copy in Arabidopsis, and (b) they had coverage consistent with single-copy status in the H. parviflora Bartl. genome skimming work. This means that a fully or mostly assembled genome is not needed to identify low-copy targets, although such resources are useful when they are available (Weitemier et al., 2014). We used GeneSeqer (Brendel et al., 2004) to identify intron-containing loci among these hits. GeneSeqer aligns transcriptome sequences to genomic sequences using several available splice-site models, evaluates putative alignments based both on sequence similarity and splice-site strength, and outputs gapped sequence alignments. In our experience, these alignments are more accurate in the context of large indels (= introns) than those based on simple sequence alignment such as BLAST. The splice-site model we used was based on Arabidopsis thaliana (L.) Heynh. Our transcriptome sequences came from a SOAP assembly of H. sanguinea (courtesy of the 1KP project, D. Soltis, and G. Wong; assembly parameters available at https://pods.iplantcollaborative.org/wiki/display/iptol/Access+to+OneKP+data+set), and the genomic data were the set of H. parviflora Velvet contigs that matched the COS marker data set. All GeneSeqer alignments were evaluated by eye to verify acceptable exon sequence identity (i.e., no obvious paralogy or spurious alignment) between H. sanguinea and H. parviflora var. saurensis. We mostly favored loci with at least two introns >100 bp long for locus development. A minority of chosen loci had a single longer intron, and two loci were included that were entirely exonic but appeared unusually variable based on alignment. We designed a 278-locus panel, synthesized by MYcroarray (Ann Arbor, Michigan) as 8634 120-bp oligomers tiled at 3× coverage of the target loci. The mean locus length was 1362 bp (range 555–3672 bp), for a total targeted length of 378,553 bp.
Library preparation
Genomic DNAs were extracted as described previously (Folk and Freudenstein, 2014). DNA was quantified with a Qubit BR-assay (Life Technologies, Carlsbad, California), and quality was evaluated using a Nanodrop 2000 (Thermo Scientific, Waltham, Massachusetts, USA). Problematic DNAs were cleaned with a QIAquick kit (QIAGEN, Valencia, California, USA); any DNAs too dilute for library preparation were concentrated by vacuum centrifugation. One microgram of clean genomic DNA in 60 μL was sonicated using a Covaris machine (model S220; Covaris, Woburn, Massachusetts, USA), aiming for fragment size of 500–700 bp. Libraries were prepared with 55.5 μL of sonicated DNA, using a NEBNext Ultra kit (New England Biolabs, Ipswich, Massachusetts, USA), following the manufacturer’s protocols with the following modifications: size selection aimed for a 500–700-bp range (i.e., 30 μL AM XP beads [Beckman Coulter, Brea, California, USA] for the first size-selection step, and 15 μL for the second step), and PCR amplification mostly used six cycles (eight cycles for the herbarium specimen). Libraries were barcoded using NEBNext Multiplex oligos (New England Biolabs). Library quality was verified using an Agilent Bioanalyzer (Agilent Technologies, Santa Clara, California, USA), and libraries were again quantified by Qubit. Any libraries that were too dilute for sequence hybridization were concentrated in a vacuum centrifuge.
Probe-based target enrichment
Verified libraries were used in a MYcroarray MyBaits in-solution hybridization protocol using our custom phylogenetic locus panel, following manufacturer protocols (version 2) with the following modifications: eight libraries were pooled per MyBaits reaction (60 ng each for 480 ng total input DNA in 6 μL) to reduce reagent cost and required input DNA, and hybridization was conducted for 36 h. PCR conditions used an annealing temperature of 60°C, an elongation time of 45 s, 15 total cycles, and 10 μL of template. Target-enriched library pools were verified for size distribution on an Agilent Bioanalyzer, and quantified with an Illumina library quantification kit (KAPA, Wilmington, Massachusetts, USA). Because adapter dimers were problematic for one of the pools, all pools were cleaned following the paramagnetic bead-based cleaning procedure in the NEBNext protocol but with 0.8 volumes of AMPure beads to 1 volume of enriched library. For all target enrichment pools, efficiency of enrichment was evaluated using two relative qPCR assays (qPCR primers, thermocycler profile, and other information in Appendix S1 (192.9KB, pdf) ). Enriched samples that amplified at least 10 cycles earlier than unenriched samples (>1000-fold enrichment assuming 100% primer efficiency) were considered successful.
Target-enriched library pools were again pooled and diluted for sequencing on an Illumina MiSeq (W. Harry Feinstone Center for Genomic Research, University of Memphis, Tennessee; 500-cycle kit, Illumina V2 chemistry). The MiSeq controller software was set to trim adapters and deconvolute barcodes; the raw output has been uploaded to the National Center for Biotechnology Information (NCBI) Sequence Read Archive (accession SRP057104). Read end trimming was performed in Trimmomatic 0.33 (Bolger et al., 2014) with a sliding window of 20 bp and a quality score of Q20 or greater.
Low copy marker sequence assembly
Assembly of data for each species reused as a reference the initial 278 contigs of H. parviflora var. saurensis that were used for probe design. We reasoned that contigs that were sufficient for designing efficient enrichment probes should also be able to inform sequence assembly, avoiding the need for computationally demanding and often unsatisfactory de novo methods. We mapped reads to the 278 references using the relatively indel-tolerant BWA program (v. 0.7.12; Li and Durbin, 2009; Li, 2013), with the default settings for paired-end data; finally, the resultant contigs were imported into Geneious. Consensus sequences were extracted from these contigs in Geneious using default settings, including trimming the consensus to the length of the reference sequence. Each locus was aligned individually using MAFFT 7.017 (Katoh et al., 2009) with default settings. One of the loci (labeled Locus 4 in the probe panel, wherein loci are numbered in order of descending length; data available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.4cn66 [Folk et al., 2015]) was found to yield only partial sequences from many taxa, resulting in large amounts of missing data (see Results); this has been excluded from the analyses, leaving 277 effectively enriched loci.
Off-target sequence assembly
We repeated the reference-mapping in BWA with mitochondrial and chloroplastic references to attempt to construct chloroplast genomes and mitochondrial genomes from off-target reads. For the chloroplast assembly, we used fully assembled plastomes from H. parviflora var. saurensis, H. parishii Rydb. (below), and H. sanguinea (M. Moore, unpublished data). The closest of these three references to each sample based on a preliminary Sanger chloroplast phylogeny (unpublished data) was used for reference-mapping to assist in assembling long, divergent intergenic regions. Development of a third plastome reference was necessary for the divergent intergenic regions of H. parishii, H. elegans Abrams, and H. abramsii Rydb.; this was constructed by mapping the H. parishii reads to the H. parviflora var. saurensis plastome in BWA and repeating the assembly refinement methods used for the H. parviflora plastome assembly. Mitochondrial assembly used the fully assembled reference from H. parviflora var. saurensis. Because we desired purely mitochondrial signal for the mitochondrial phylogenetic analysis, this genome was blasted against the H. parviflora chloroplast genome and any hits were deleted from the mitochondrial assembly reference. Whole chloroplast genomes were aligned in MAFFT as with the single-copy markers; however, this was computationally infeasible for the much longer mitochondrial sequences. For these we used Mauve, setting an assumption of collinear genomes (which was enforced for our contigs by the read-mapping method) but otherwise using default settings.
Phylogenetic analysis
Because chloroplast capture is a well-known phenomenon in the genus Heuchera (Soltis et al., 1991; Soltis and Kuzoff, 1995; Folk, Mandel, and Freudenstein, unpublished data), and organellar genomes in general are thought to be more prone to displaying phylogenetic discordance caused by hybridization (Rieseberg and Soltis, 1991), we avoided including organellar genes in all multilocus analyses and instead present these separately. We performed a concatenated analysis of the 277 low-copy nuclear loci in RAxML 8.1.3 (Stamatakis, 2006), using an unpartitioned GTR-GAMMA model and with support quantified by 5000 bootstrap replicates (matrices and tree files for the analyses of this paper available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.4cn66 [Folk et al., 2015]).
To examine the relative phylogenetic signal from exonic and intronic portions of the sequences, we performed an exon-only analysis and an intron-only analysis by alternatively excluding intron and exon sites, respectively. Approximately two thirds of our target sequence was intronic; hence analyzing this in its entirety would be an unfair comparison with the exonic matrix. Therefore, we arbitrarily excluded the last 49.3% of the intron matrix (introns of approximately loci 88–277) to yield a matrix of equivalent length to the exon matrix. Each of these two matrices was analyzed in RAxML with an unpartitioned GTR-GAMMA model, with 1000 bootstrap replicates.
The 277 gene alignments were also analyzed individually in RAxML to infer gene trees, with the GTR-GAMMA model and 1000 bootstrap replicates. The optimal trees and bootstrap samples from each gene tree were analyzed using a gene tree–based coalescent approach in MP-EST (Liu et al., 2010) and STAR (Liu et al., 2009) using the STRAW server (Shaw et al., 2013).
Mitella pentandra Hook. was used as the outgroup for the nuclear analysis based on the results in Folk and Freudenstein (2014). Rooting is not as straightforward for the chloroplast data set because Heuchera is known to be polyphyletic for chloroplast markers due to frequent hybridization with closely related genera such as Tiarella L. (Soltis et al., 1991; Folk, Mandel, and Freudenstein, unpublished data). If a supposed outgroup had captured a Heuchera chloroplast genome, this would result in incorrectly rooting the tree within Heuchera. While a mitochondrial analysis has never been undertaken for the Heuchera group of genera, these concerns could also apply to the mitochondrial data. To address these concerns, we repeated the chloroplast and mitochondrial read-mapping approach for publicly available short read data from Saxifraga granulata L. (one individual from Meer et al., 2014) to develop an appropriate outgroup sequence well outside the Heuchera group of genera.
The low-coverage of mitochondrial DNA and the greater divergence from the reference resulted in a greater amount of missing data than in other data sets, especially in intergenic regions; this was addressed by deleting all columns with ≥75% missing data (a trial was also made of deleting columns with 50% missing data, but this resulted in lowered support values on an identical topology; results not shown). Otherwise, the same phylogenetic analysis parameters from the low-copy nuclear markers were used for the mitochondrial and chloroplast data sets.
Any differences in support values between this analysis and the previous phylogenetic context of Folk and Freudenstein (2014) could be caused by reduced taxon sampling rather than a larger data set, as analyzing fewer taxa reduces the tree search space and may lengthen short branches that would have been broken up by intermediate taxa. To evaluate this possibility, we reanalyzed a reduced version of the earlier matrix, using only DNA characters and eliminating all taxa not sampled in this study. Heuchera acutifolia could not be sequenced for most markers in the earlier study, so we substituted the close relative H. longipetala Moc. ex Ser. For comparability, the matrix was run with an unpartitioned GTR-GAMMA model with 1000 bootstrap replicates.
Enrichment statistics
Percent on-target statistics (Table 1) were calculated by mapping reads to the reference loci using the Geneious native assembler (low sensitivity settings) and dividing the number of mapped reads by the total number of reads per each accession; the same method was used to determine read percentages for the chloroplast and mitochondrial genomes using the mitochondrial and chloroplast references. Other assembly statistics (Table 1) were derived from the BWA assemblies used in downstream analyses, but also calculated in Geneious. Missing data, pairwise identity, and other alignment statistics (Table 2) were derived in Geneious from the MAFFT alignments.
Table 1.
Statistics for the target enrichment experiment.a
| Accession | Percent on-target | Mean (±SD) of low-copy locus coverage | Mean low-copy locus completeness (%)b | Mean exon completeness (%)c | Mean intron completeness (%)c | Percent chloroplast reads | Chloroplast coverage | Chloroplast completeness (%) | Percent mitochondrial reads | Mitochondrial coveraged | Mitochondrial completeness (%) |
| Heuchera abramsii | 54.7 | 173.5× (±75.4×) | 99.8 | 100.0 | 99.6 | 7.8 | 31.8× | 98.3 | 1.0 | 4.5× | 44.0 |
| H. acutifolia (herbarium specimen) | 44.5 | 206.0× (±76.5×) | 99.8 | 99.9 | 99.3 | 3.8 | 22.0× | 95.3 | 1.5 | 7.2× | 66.3 |
| H. americana var. americana | 58.7 | 178.9× (±52.5×) | 99.9 | 99.9 | 99.5 | 3.7 | 14.8× | 96.6 | 1.1 | 3.7× | 52.7 |
| H. elegans | 54.8 | 464.6× (±136.5×) | 99.9 | 100.0 | 99.9 | 3.4 | 38.6× | 99.7 | 0.4 | 4.7× | 47.1 |
| H. grossulariifolia var. grossulariifolia | 59.6 | 262.3× (±70.3×) | 100.0 | 100.0 | 99.7 | 6.4 | 36.9× | 99.4 | 0.9 | 4.5× | 58.5 |
| H. missouriensis | 60.5 | 586.1× (±112.1×) | 100.0 | 99.8 | 100.0 | 4.2 | 63.4× | 99.8 | 0.6 | 4.9× | 81.3 |
| H. parishii | 47.3 | 922.9× (±236.3×) | 99.9 | 100.0 | 100.0 | 3.8 | 99.8× | 99.8 | 0.4 | 8.2× | 64.2 |
| H. parviflora var. parviflora | 60.6 | 1225.7× (±242.9×) | 100.0 | 100.0 | 100.0 | 6.7 | 220.2× | 99.7 | 1.1 | 17.8× | 85.3 |
| H. parvifolia var. nivalis | 52.8 | 714.9× (±168.1×) | 100.0 | 100.0 | 100.0 | 3.0 | 51.8× | 99.7 | 1.0 | 9.4× | 84.7 |
| H. puberula | 55.0 | 440.4× (±93.5×) | 100.0 | 100.0 | 100.0 | 4.0 | 47.3× | 99.8 | 0.9 | 6.6× | 73.1 |
| H. pulchella | 47.8 | 1303.8× (±254.1×) | 99.9 | 100.0 | 100.0 | 5.5 | 219.3× | 99.9 | 0.7 | 13.1× | 90.8 |
| H. rubescens var. versicolor | 55.8 | 321.3× (±79.6×) | 99.9 | 100.0 | 99.9 | 2.4 | 17.6× | 99.5 | 0.4 | 3.5× | 42.6 |
| H. villosa var. villosa | 62.6 | 361.9× (±76.6×) | 100.0 | 100.0 | 99.9 | 2.9 | 24.7× | 99.1 | 0.6 | 3.5× | 61.4 |
| H. wootonii | 55.7 | 275.9× (±72.6×) | 99.8 | 99.9 | 99.6 | 2.8 | 17.9× | 99.1 | 0.9 | 4.8× | 60.2 |
| Mitella pentandra | 49.8 | 238.9× (±91.7×) | 99.6 | 99.6 | 98.5 | 7.2 | 41.3× | 98.6 | 1.6 | 6.6× | 70.2 |
For all calculations, locus four has been omitted.
Locus completeness is based on coverage of the reference sequences.
Region completeness figures are based on the percent of the H. parviflora reference to which reads could be mapped.
Coverage of the mitochondrion only counts areas where reads could be mapped. If this calculation included the whole mitochondrion, it would be unfairly down-weighted by long regions (up to tens of thousands of bases) of uncertain origin that are not shared across species.
Table 2.
Statistics for the phylogenetic alignments of low-copy nuclear loci.
| Sequence statistic | Entire experiment | Exons only | Introns only |
| Percent pairwise identity (pairwise divergence) | 96.4% (3.6%) | 98.8% (1.2%) | 95.2% (4.8%) |
| Number (percent) parsimony informative characters | 17,691 (4.6%) | 1830 (1.5%) | 15,861 (6.1%) |
| Percent invariant characters | 82.90% | 93.40% | 77.80% |
| Undetermined sequence (N or ?) | 0.20% | 0.10% | 0.30% |
| Gap characters (-) | 3.00% | 0.10% | 4.30% |
| Overall alignment length | 387,941 bp | 126,540 bp | 261,401 bp |
| Average coverage per locus | 511.8× | ||
| Average locus length | 1362 bp | ||
| Percent on-target reads for H. parviflora var. saurensis (unenriched genome skimming) | 0.06% |
RESULTS
Target enrichment
Our target enrichment methodology resulted in between 45% and 63% on-target sequences. This range is relatively high for interspecies target enrichment (e.g., compare Weitemier et al., 2014), and may result from the particularly low divergence among species of Heuchera, causing high complementarity between probes and target sequences. This resulted in relatively high coverage numbers for the study loci; although coverage numbers were variable, all samples had greater than 100× average coverage (Table 1). Low coverage for target loci was extremely rare, other than for the excluded locus (see Methods). This locus appears to be a pseudogene; no well-matched short reads of this locus were obtained for about half of the samples.
The clear explanation for the difference in coverage between samples is uneven pooling, rather than on-target sequence percentage, which did not vary particularly greatly between species. Hence the success of target enrichment was not markedly different between samples. Even for the outgroup Mitella pentandra, where slight signs of intron dropout are apparent (∼1% lower assembly completeness, Table 1), it remained possible to assemble the vast majority of intronic sequences.
Comparison with the proportion of on-target reads for the unenriched genome-skimming sample (0.057%, vs. average 54.7% on target for enriched individuals) showed that the fold enrichment was very high (0.547/0.00057 ∼960-fold enrichment assuming constant background prevalence of target loci across samples), consistent with >10 cycles increase in target concentration in the qPCR assays for all enriched individuals. Overall locus assembly completeness was also nearly 100% for all samples (Table 1); this applies both to exons and introns, the latter of which had negligibly lower completeness.
Percent pairwise identity calculations from the phylogenetic matrix (Table 1) show that for Heuchera species, the percent divergence of introns is fourfold that of exons. Similarly, introns had about four times the percent parsimony-informative characters. While nucleotide diversity is high in introns, as expected they are also quite rich in indel characters, >40-fold more so than exons.
Phylogenetic inference
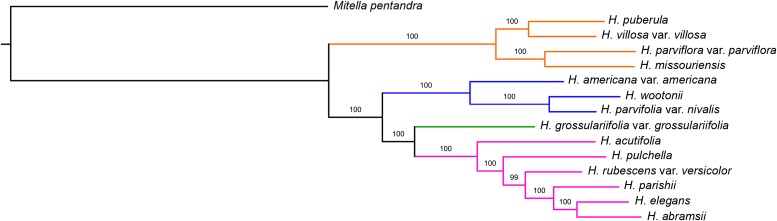
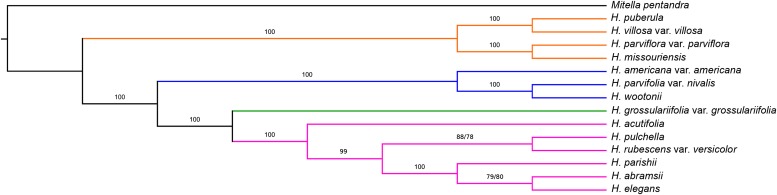
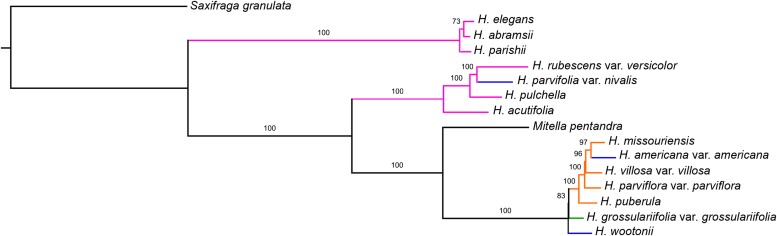
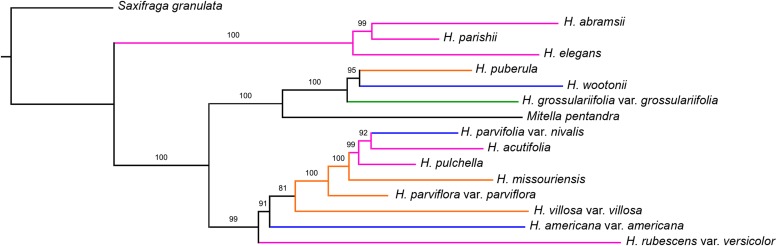
The concatenated nuclear tree (Fig. 1) was completely resolved with strong support on all nodes (99% on one node in sect. Rhodoheuchera concerning the placement of H. rubescens Torr. var. versicolor (Greene) M. G. Stewart; otherwise 100%). Coalescent analyses in MP-EST and STAR (Fig. 2) likewise had primarily high support values. There was only one point of discord between these methods: coalescent methods found H. pulchella Wooton & Standl. and H. rubescens var. versicolor as sister to each other, whereas concatenation inferred these as successively sister to H. parishii, H. elegans, and H. abramsii. The phylogenetic analyses of organellar data (chloroplast, Fig. 3; mitochondrial, Fig. 4) likewise resulted in high support values, although these relationships differed greatly from those suggested by nuclear data.
Fig. 1.
Concatenated maximum likelihood (ML) tree based on 277 low-copy nuclear loci; branch lengths in the figure represent ML branch length estimates. Coloring of branches represents the taxonomic sections recognized in Folk and Freudenstein (2014): orange = sect. Holochloa; blue = sect. Heuchera; green = sect. Bracteatae; magenta = sect. Rhodoheuchera. Values plotted on branches are support values based on 5000 bootstrap replicates. Varietal assignments in sections Heuchera and Rhodoheuchera reflect the taxonomy of the treatments by Rosendahl et al. (1936) and Wells (1984); taxon labels in sect. Holochloa reflect Folk and Freudenstein (in press).
Fig. 2.
Coalescent tree estimated in MP-EST. Branch labels are bootstrap supports, based on 1000 bootstrap replicates per tree. The STAR result was topologically identical; where support values differed the two values are plotted as “MP-EST proportion/STAR proportion.” Branch coloring follows the sectional taxonomy as in Fig. 1; branch lengths are not to scale.
Fig. 3.
Maximum likelihood tree of the chloroplast data set. Branch proportions, coloring, and labeling follow Fig. 1, except that the outgroup branch length is not to scale.
Fig. 4.
Maximum likelihood tree of the mitochondrial data set. Branch coloring and labeling follow Fig. 1, except that the outgroup branch length is not to scale.
Organellar reference genomes
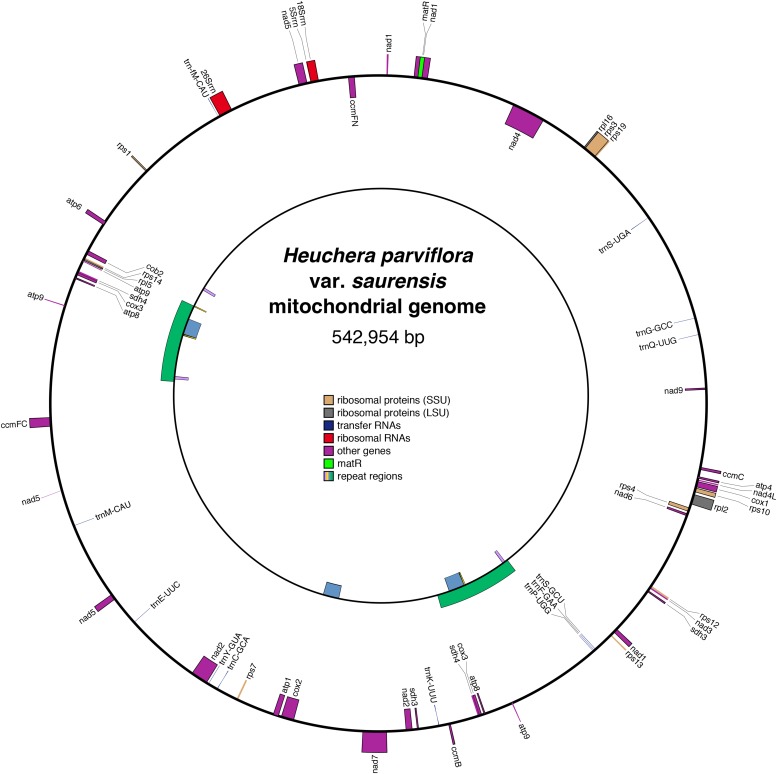
The chloroplast genome of H. parviflora var. saurensis (154,696 bp; mean coverage 1939.8×; no map shown) is conventional among angiosperms in terms of structure and size, with complete synteny shared with other members of the order. The H. parviflora var. saurensis mitochondrial genome (541,954 bp; mean coverage 141.9×, draft map in Fig. 5) is much less conserved in terms of structure, as is typical for plant mitochondrial genomes (Sloan, 2013), with only a few very short regions of synteny and only short stretches of similar sequence preserved when compared to the closest reference, V. vinifera (Appendix S2 (887.4KB, pdf) ). It has all functional mitochondrial protein-coding genes that are present in Vitis, as well as numerous interspersed sequences of chloroplast origin. BLAST searches against the Marchantia polymorpha L. mitochondrial genome showed that all conserved mitochondrial genes absent in Vitis were also absent in Heuchera. Processes of intramolecular recombination and multiple conformations of the plant mitochondrial genome are thought to be mediated by moderately long repeat regions (Alverson et al., 2011, and citations therein), of which we found four. These repeat regions were independently supported by locally higher read coverage. Unusually, the smaller three repeat regions (6428 bp, 1196 bp, 550 bp) that we found overlap with the largest repeat (32,849 bp), but with a single copy of each elsewhere in the genome. The issue of multiple genome conformations is not critical for assemblies intended for analysis with phylogenetic methods. Using these two organellar references, we were able to infer resolved phylogenies with high support values as well (chloroplast, Fig. 4; mitochondrion, Fig. 5).
Fig. 5.
Map of regions of interest in the mitochondrial genome of Heuchera parviflora var. saurensis, prepared using OrganellarGenomeDRAW (Lohse et al., 2013). The outer circle depicts protein-coding genes, rRNAs, and tRNAs; the inner circle depicts the large repeat regions. Genes annotated on the inner face of the circle indicate genes transcribed in a clockwise orientation, and genes on the outer face are transcribed counterclockwise. The orientation of the repeat regions is arbitrary; the green and blue regions are direct repeats, while the yellow and lavender regions exist both as direct repeats (where they overlap the green direct repeat) and inverted repeats (elsewhere). Annotated gene regions include both introns and exons. Plant mitochondrial genomes have several trans-spliced genes (e.g., nad5); only the cistronic portions are annotated to avoid overlap.
DISCUSSION
Low-copy locus design
Most eukaryotic nuclear genes have introns; this splice site structure is highly conserved (The Arabidopsis Initiative, 2000; Roy et al., 2003), making these regions a practically inexhaustible source of phylogenetic signal. Fully assembled genomes are not required for marker development. At minimum, however, intronic locus design for targeted sequencing requires the researcher to obtain a large number of contigs at least as long as the desired marker length, as well as transcriptomic sequences to identify splice sites. Finally, some method must be used to select putative low-copy genes, such as curated genome drafts from the study organism (de Sousa et al., 2014; Weitemier et al., 2014) or, when this information is absent, panels of loci derived from model organism genomes (Mandel et al., 2014; this study). In the latter case, if genome skimming data are available from one of the study organisms, coverage data are effective as an independent source of evidence for low-copy marker status.
Concatenated nuclear analysis
The relationships recovered were broadly congruent with results based on Sanger sequencing (Folk and Freudenstein, 2014); however, the increase in support was remarkable in several instances. The improvement in support over earlier work was most remarkable for section Rhodoheuchera (Fig. 1, marked in magenta), the largest in the genus (∼17 species). In Folk and Freudenstein (2014), this section was the most problematic group for phylogenetic inference; there was weak or no support for all relationships in the section, and at best moderate support for the monophyly of the section as a whole. The new data set confidently confirms the monophyly of sect. Rhodoheuchera and resolves relationships in this group.
The analysis of the reduced Folk and Freudenstein (2014) matrix (Appendix S3 (67.7KB, pdf) ) showed that higher support values were due to a mixture of reduced taxon sampling and more informative data. The average nodal bootstrap (BS) proportion was 100% for the entire phylogenomic data set and 78.4% for the Sanger data set; several nodes in the Sanger data set were higher than in earlier work. Nevertheless, the improvement caused by greater locus sampling is apparent; the addition of further taxa would inflate the tree search space, likely causing a greater disparity between the two data sets. It is important to note that the Sanger tree was incongruent in several places with the phylogenomic data set; particularly, section Heuchera (blue, Appendix S3 (67.7KB, pdf) ) is sister to the rest of the genus (BS 76). Likewise, the positions of H. rubescens var. versicolor (BS 45) and H. parviflora var. parviflora (BS 99) differed. The position of section Heuchera is congruent with the earlier study, while the positions of the other two were equivocal. The relatively high bootstraps for two of these anomalous Sanger relationships is unlikely to have been caused by reduced taxon sampling; it is more likely that the smaller data set inadequately sampled the genome for conflicting phylogenetic signals for the positions of these taxa.
Intron- and exon-only phylogenetic analyses (Appendices S4 (69.4KB, pdf) , S5 (67.9KB, pdf) ) revealed further differences in these data types. Support values for the two analyses were only somewhat different (the intron-only analysis had 96.8% average nodal support; the exon-only analysis had 94.8% average support). However, only the intron-only analysis was congruent with the full data set result. For the exon-only analysis, H. rubescens var. versicolor and H. acutifolia exchanged topological positions (BS 78). Because the full analysis also addressed gene tree heterogeneity through coalescent analysis, the exon-only result does not appear to be optimal.
Coalescent nuclear analysis
While we are aware of the issues associated with using long genetic loci with multispecies coalescent methods and the importance of incorporating gene tree estimation error (Gatesy and Springer, 2014), nevertheless we contend that a coalescent analysis retains heuristic value, particularly in concert with the more standard concatenation methods. Especially when only well-supported nodes are considered, for this data set the difference between coalescent and concatenated estimates of the species tree is not particularly great. It is interesting that the only point of discord between concatenated and coalescent approaches was one of only two nodes in the coalescent analysis below 95% (= strong support; however, at 88% [MP-EST] or 78% [STAR] it is still moderate); Folk and Freudenstein (2014) also noted that points of discord between coalescence and concatenation estimates tend to be associated with lower nodal support in the coalescent tree. On the other hand, MP-EST and STAR were remarkably consistent with one another.
Chloroplast phylogeny
Heuchera is known as a particularly dramatic example of chloroplast capture (Soltis et al., 1991), although in the time that has passed since this system was examined it has become possible to sequence essentially entire chloroplast genomes to explore this signal further. In the current analysis (Fig. 4), the chloroplast genome assemblies are practically complete; the few missing regions tend to correspond to long A-T repeats and other types of microsatellite-like regions (total aligned length 158,903 bp; 1.8% missing data). This study serves as the first sequence-based confirmation of the discord between nuclear and chloroplast phylogenies (see Soltis et al., 1991; Soltis and Kuzoff, 1995) and uncovers new discord beyond what has previously been observed. The clade of Heuchera in this analysis that resolves as sister to Mitella pentandra corresponds to a clade hypothesized to have captured the Tiarella plastome (Soltis et al., 1991), while the clade consisting of H. parvifolia, H. acutifolia, and H. pulchella corresponds to the clade hypothesized to possess the ancestral plastid genome. However, the position of H. parishii, H. elegans, and H. abramsii (species unsampled in the earlier study) as a clade sister to the rest of the Heuchera group of genera represents a new lineage not uncovered previously.
Mitochondrial phylogeny
We acknowledge that, due to the lower coverage of the mitochondrial data in many samples (mostly caused by the lower background prevalence and larger size of mitochondrial genomes as compared to plastomes), our mitochondrial phylogeny should be viewed as experimental. The lack of conservation of basic structural features and even large stretches of sequence further hinders assembly. Nevertheless, it is remarkable that our mitochondrial analysis (total aligned length 421,260 bp; 43.9% missing data) resulted in a resolved topology, largely with strong support values, in a genus for which mitochondrial phylogenetics has never been performed.
The mitochondrial phylogeny (Fig. 5) shows a similar degree of conflict with the nuclear topology as does the chloroplast tree. In fact, there are some parallel points of conflict between the data sets. Particularly, the position of H. parishii, H. elegans, and H. abramsii as sister to all other members of the Heuchera group of genera is exactly congruent with the chloroplast analysis, although the topology within this clade differs somewhat. Such mitochondrial skimming results have broad relevance for plant systematists. Mitochondrial markers have been under-sampled in plant phylogenetics as compared to nuclear and chloroplast markers; in plants, mitochondrial loci have seen use mainly at higher levels (e.g., Chaw et al., 2000; Jian et al., 2008), and among conifer species (Gugerli et al., 2001), and only occasionally studied alone to infer mitochondrial trees. Some work has successfully used mitochondrial skimming (Straub et al., 2012; Ripma et al., 2014, Govindarajulu et al., 2015), but this remains less common than the use of chloroplast data. Mitochondrial capture is well-known in animals (Good et al., 2008, and citations therein); hence in plant groups, particularly those that are believed to have undergone chloroplast capture, mitochondrial phylogenies promise to further and independently resolve past hybridization events (see also Govindarajulu et al., 2015). The current paucity of good reference mitochondrial genomes among angiosperms may be among the major setbacks preventing resolved mitochondrial phylogenetic hypotheses in many plant groups. Given their unusual patterns of evolution, it will be interesting to explore whether mitochondrial phylogenies show capture events as frequently as has been observed for chloroplast markers.
Conclusions
Even given large phylogenomic data sets, sequencing methods and marker choice must be no less carefully optimized for particular research questions. Recent radiations straddle the boundary of phylogenetic and population-level processes, lending themselves to targeted enrichment of intronic markers. For these systems, where coalescent branch lengths are short and barriers to gene flow often poorly developed, gene tree discord is likely to be prevalent. Examining conflicting histories necessitates sequences long enough to infer resolved gene trees. The ability to assemble large amounts of off-target organellar data with such methods further results in a robust sampling of alternative genomic histories. Likewise, sequence divergence will usually be low in recent radiations, with the result that targeting more conserved portions of the genome may waste sequencing effort. Compounding these difficulties for any inferential method is the effect of missing data; we have shown that this method results in essentially no missing data for targeted nuclear markers and chloroplast data (in contrast to a typical RNA-seq or RAD-seq experiment; see also Weitemier et al., 2014).
In the increasingly data-rich field of systematics, phylogenomic conflict is becoming the rule rather than the exception, with the result that reporting a single total-evidence tree without examining the underlying data is becoming less satisfactory. As systematists grapple with gene tree heterogeneity in the most difficult systems, targeted methods optimized for gene tree resolution are poised to address all of these concerns. Intron-containing low-copy sequence markers, long-favored for Sanger sequencing (Folk and Freudenstein, 2014, and citations therein), will be increasingly important in genera and species complexes for a robust sampling not only for a total-evidence phylogeny, but more critically for individual gene histories.
Supplementary Material
Appendix 1.
Voucher information for accessions used in this study.
| Species | Herbarium voucher no. (Herbariuma) | Collection locality | Geographic coordinates |
| Heuchera abramsii Rydb. | Folk 45 (OS) | Mount Baldy, CA, USA | 34°17.423′N, 117°38.738′W |
| H. acutifolia Rose (herbarium specimen) | Matus 14560 (TEX) | Rancho Poza, VER, Mexico | 20°32′10.38″N, 98°28′51.58″W |
| H. americana L. var. americana | Folk 70 (OS) | Berry Road, Wayne National Forest, OH, USA | 38°36′9.11″N, 82°20′22.16″W |
| H. elegans Abrams | Folk 44 (OS) | Talus above road to Crystal Lake, 0.5 mi. from Hwy. 39, CA, USA | 43°18.940′N, 117°50.537′W |
| H. grossulariifolia Rydb. var. grossulariifolia (tetraploid) | Folk 160 (OS) | Cliff above roadside parking lot outside of Riggins, ID, USA | 45°24′40″N, 116°19′35″W |
| H. missouriensis Rosend. | Folk I-69 (OS) | Little Grand Canyon near Carbondale, IL, USA | 37°41′4.46″N, 89°24′38.92″W |
| H. parishii Rydb. | Folk 43 (OS) | Sugarloaf Mountain, CA, USA | 34°12.003′N, 116°47.621′W |
| H. parviflora Bartl. var. parviflora | Folk 72 (OS) | Natural Bridge State Park, KY, USA | 37°46′26.33″N, 83°40′51.39″W |
| H. parvifolia var. nivalis (Rosend., Butters & Lakela) Á. Löve, D. Löve & B. M. Kapoor | Folk 55 (OS) | Along Rock Creek Hills Road, Lost Park, CO, USA | 39°20′59″N, 105°41′25″W |
| H. puberula Mack. & Bush | Folk 69 (OS) | Pea Vine Road near Big Springs, Ozark National Riverways, MO, USA | 36.94794°N, 90.99215°W |
| H. pulchella Wooton & Standl. | Folk 20 (OS) | Sandia Crest, NM, USA | 35°12.662′N, 106°27.029′W |
| H. rubescens Torr. var. versicolor (Greene) M. G. Stewart | Folk 26 (OS) | Outcrop above Hwy. 159, NM, USA | 33°22′14″N, 108°41′34″W |
| H. villosa Michx. var. villosa | Folk I-1 (OS) | Devil’s Courthouse, NC, USA | 35°18′9.44″N, 82°53′44.39″W |
| H. wootonii Rydb. | Folk 22 (OS) | Dry bank, Capitan Peak, NM, USA | 33°36′45″N, 105°14′45″W |
| Mitella pentandra Hook. | Folk 128 (OS) | Pacific Crest Trail access from Hwy. 90, WA, USA | 47 25′40″N, 121 24′48″W |
OS = Ohio State University Herbarium; TEX = University of Texas at Austin Herbarium.
LITERATURE CITED
- Alverson A. J., Zhuo S., Rice D. W., Sloan D. B., Palmer J. D. 2011. The mitochondrial genome of the legume Vigna radiata and the analysis of recombination across short mitochondrial repeats. PLoS One 6: e16404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Arabidopsis Initiative. 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815. [DOI] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., Usadel B. 2014. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendel V., Xing L., Zhu W. 2004. Gene structure prediction from consensus spliced alignment of multiple ESTs matching the same genomic locus. Bioinformatics 20: 1157–1169. [DOI] [PubMed] [Google Scholar]
- Cao J., Schneeberger K., Ossowski S., Günther T., Bender S., Fitz J., Koenig D., et al. 2011. Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nature Genetics 43: 956–963. [DOI] [PubMed] [Google Scholar]
- Chaw S.-M., Parkinson C. L., Cheng Y., Vincent T. M., Palmer J. D. 2000. Seed plant phylogeny inferred from all three plant genomes: Monophyly of extant gymnosperms and origin of Gnetales from conifers. Proceedings of the National Academy of Sciences, USA 97: 4086–4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronn R., Knaus B. J., Liston A., Maughan P. J., Parks M., Syring J. V., Udall J. 2012. Targeted enrichment strategies for next-generation plant biology. American Journal of Botany 99: 291–311. [DOI] [PubMed] [Google Scholar]
- Darling A. C. E. 2004. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Research 14: 1394–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sousa F., Bertrand Y. J. K., Nylinder S., Oxelman B., Eriksson J. S., Pfeil B. E. 2014. Phylogenetic properties of 50 nuclear loci in Medicago (Leguminosae) generated using multiplexed sequence capture and next-generation sequencing. PLoS One 9: e109704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J.-B., Drew B. T., Mavrodiev E. V., Gitzendanner M. A., Soltis P. S., Soltis D. E. 2015. Phylogeny, divergence times, and historical biogeography of the angiosperm family Saxifragaceae. Molecular Phylogenetics and Evolution 83: 86–98. [DOI] [PubMed] [Google Scholar]
- Folk R. A., Freudenstein J. V. 2014. Phylogenetic relationships and character evolution in Heuchera (Saxifragaceae) on the basis of multiple nuclear loci. American Journal of Botany 101: 1532–1550. [DOI] [PubMed] [Google Scholar]
- Folk R. A., Mandel J. R., Freudenstein J. V. 2015. Data from: A protocol for targeted enrichment of intron-containing sequence markers for recent radiations: A phylogenomic example from Heuchera (Saxifragaceae). Dryad Digital Repository. http://dx.doi.org/10.5061/dryad.4cn66 [DOI] [PMC free article] [PubMed]
- Gatesy J., Springer M. S. 2014. Phylogenetic analysis at deep timescales: Unreliable gene trees, bypassed hidden support, and the coalescence/concatalescence conundrum. Molecular Phylogenetics and Evolution 80: 231–266. [DOI] [PubMed] [Google Scholar]
- Gnirke A., Melnikov A., Maguire J., Rogov P., LeProust E. M., Brockman W., Fennell T., et al. 2009. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nature Biotechnology 27: 182–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godsoe W., Larson M. A., Glennon K. L., Segraves K. A. 2013. Polyploidization in Heuchera cylindrica (Saxifragaceae) did not result in a shift in climatic requirements. American Journal of Botany 100: 496–508. [DOI] [PubMed] [Google Scholar]
- Good J. M., Hird S. M., Reid N. M., Demboski J. R., Steppan S. J., Martin-Nims T. R., Sullivan J. 2008. Ancient hybridization and mitochondrial capture between two species of chipmunks. Molecular Ecology 17: 1313–1327. [DOI] [PubMed] [Google Scholar]
- Goremykin V. V., Salamini F., Velasco R., Viola R. 2008. Mitochondrial DNA of Vitis vinifera and the issue of rampant horizontal gene transfer. Molecular Biology and Evolution 26: 99–110. [DOI] [PubMed] [Google Scholar]
- Govindarajulu R., Parks M., Tennessen J. A., Liston A., Ashman T.-L. 2015. Comparison of nuclear, plastid, and mitochondrial phylogenies and the origin of wild octaploid strawberry species. American Journal of Botany 102: 544–554. [DOI] [PubMed] [Google Scholar]
- Gugerli F., Senn J., Anzidei M., Madaghiele A., Buchler U., Sperisen C., Vendramin G. G. 2001. Chloroplast microsatellites and mitochondrial nad1 intron 2 sequences indicate congruent phylogenetic relationships among Swiss stone pine (Pinus cembra), Siberian stone pine (Pinus sibirica), and Siberian dwarf pine (Pinus pumila). Molecular Ecology 10: 1489–1497. [DOI] [PubMed] [Google Scholar]
- Jian S., Soltis P. S., Gitzendanner M. A., Moore M. J., Li R., Hendry T. A., Qiu Y.-L., et al. 2008. Resolving an ancient, rapid radiation in Saxifragales. Systematic Biology 57: 38–57. [DOI] [PubMed] [Google Scholar]
- Katoh K., Asimenos G., Toh H. 2009. Multiple alignment of DNA sequences with MAFFT. In D. Posada , Methods in molecular biology, vol. 537: Bioinformatics for DNA sequence analysis, 39–64. Humana Press, Totowa, New Jersey, USA. [DOI] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. ArXiv arXiv:1303.3997.
- Li H., Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Yu L., Pearl D. K., Edwards S. V. 2009. Estimating species phylogenies using coalescence times among sequences. Systematic Biology 58: 468–477. [DOI] [PubMed] [Google Scholar]
- Liu L., Yu L., Edwards S. V. 2010. A maximum pseudo-likelihood approach for estimating species trees under the coalescent model. BMC Evolutionary Biology 10: 302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M., Drechsel O., Kahlau S., Bock R. 2013. OrganellarGenomeDRAW—A suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Research 41: W575–W581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R., Liu B., Xie Y., Li Z., Huang W., Yuan J., He G., et al. 2012. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. GigaScience 1: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel J. R., Dikow R. B., Funk V. A., Masalia R. R., Staton S. E., Kozik A., Michelmore R. W., et al. 2014. A target enrichment method for gathering phylogenetic information from hundreds of loci: An example from the Compositae. Applications in Plant Sciences 2: 1300085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack J. E., Hird S. M., Zellmer A. J., Carstens B. C., Brumfield R. T. 2013. Applications of next-generation sequencing to phylogeography and phylogenetics. Molecular Ecology 66: 526–538. [DOI] [PubMed] [Google Scholar]
- Meer S. V. D., Houdt J. K. J. V., Maes G. E., Hellemans B., Jacquemyn H. 2014. Microsatellite primers for the gynodioecious grassland perennial Saxifraga granulata (Saxifragaceae). Applications in Plant Sciences 2: 1400040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg L. H., Soltis D. E. 1991. Phylogenetic consequences of cytoplasmic gene flow in plants. Evolutionary Trends in Plants 5: 65–84. [Google Scholar]
- Ripma L. A., Simpson M. G., Hasenstab-Lehman K. 2014. Geneious! Simplified genome skimming methods for phylogenetic systematic studies: A case study in Oreocarya (Boraginaceae). Applications in Plant Sciences 2: 1400062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosendahl C. O., Butters F. K., Lakela O. 1936. A monograph on the genus Heuchera. University of Minnesota Press, Minneapolis, Minnesota, USA. [Google Scholar]
- Roy S. W., Federov A., Gilbert W. 2003. Large-scale comparison of intron positions in mammalian genes shows intron loss but no gain. Proceedings of the National Academy of Sciences, USA 100: 7158–7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhfel B. R., Gitzendanner M. A., Soltis P. S., Soltis D. E., Burleigh J. G. 2014. From algae to angiosperms—Inferring the phylogeny of green plants (Viridiplantae) from 360 plastid genomes. BMC Evolutionary Biology 14: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang T. 2002. Utility of low-copy nuclear gene sequences in plant phylogenetics. Critical Reviews in Biochemistry and Molecular Biology 37: 121–147. [DOI] [PubMed] [Google Scholar]
- Shaw T. I., Ruan Z., Glenn T. C., Liu L. 2013. STRAW: Species TRee Analysis Web server. Nucleic Acids Research 41: W238–W241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan D. B. 2013. One ring to rule them all? Genome sequencing provides new insights into the ‘master circle’ model of plant mitochondrial DNA structure. New Phytologist 200: 978–985. [DOI] [PubMed] [Google Scholar]
- Soltis D., Soltis P., Collier T. 1991. Chloroplast DNA variation within and among genera of the Heuchera group (Saxifragaceae): Evidence for chloroplast transfer and paraphyly. American Journal of Botany 78: 1091–1112. [Google Scholar]
- Soltis D., Kuzoff R. 1995. Discordance between nuclear and chloroplast phylogenies in the Heuchera group (Saxifragaceae). Evolution 49: 727–742. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: Maximum likelihood–based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- Straub S. C. K., Parks M., Weitemier K., Fishbein M., Cronn R. C., Liston A. 2012. Navigating the tip of the genomic iceberg: Next-generation sequencing for plant systematics. American Journal of Botany 99: 349–364. [DOI] [PubMed] [Google Scholar]
- Weitemier K., Straub S. C. K., Cronn R. C., Fishbein M., Schmickl R., McDonnell A., Liston A. 2014. Hyb-Seq: Combining target enrichment and genome skimming for plant phylogenomics. Applications in Plant Sciences 2: 1400042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells E. 1984. A revision of the genus Heuchera (Saxifragaceae) in eastern North America. Systematic Botany Monographs 3: 45–121. [Google Scholar]
- Zerbino D. R., Birney E. 2008. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Research 18: 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.