Figure 3.
Quantitative Proteomics Analysis of Host Protein Fatty Acylation During HSV Infection in RPE-1 Cells
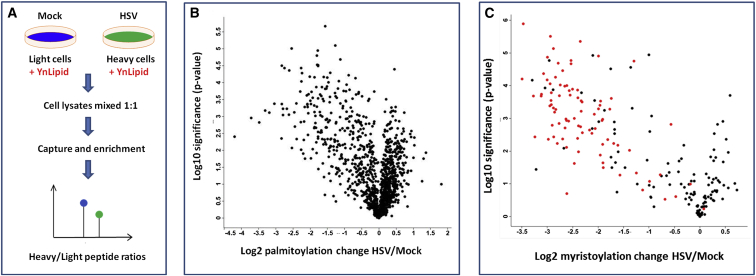
(A) SILAC-based quantitative proteomics workflow.
(B) Virus-induced changes to protein palmitoylation (n = 4) plotted against statistical significance of the ratio measured.
(C) Virus-induced changes to protein myristoylation (n = 4) plotted against statistical significance. Black, proteins with myristoylation requirement (N-terminal Gly); red, validated NMT substrates (Broncel et al., 2015; Thinon et al., 2014).
In (B) and (C) each data point represents a protein or a protein group (Cox et al., 2011).