FIG. 1.
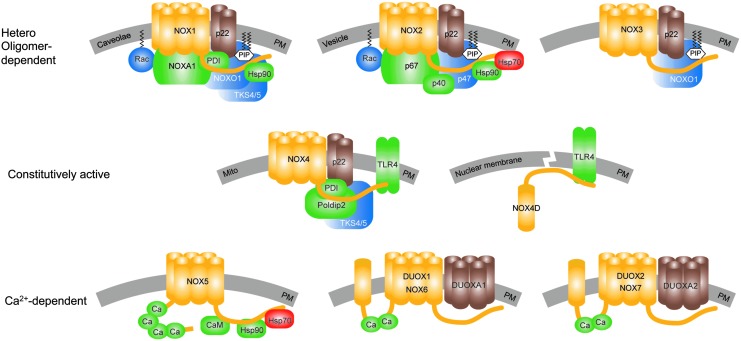
The NADPH oxidase enzyme family. All NOX isoforms (yellow) are membrane proteins that are localized in the PM or cellular compartments' membranes (gray). Stabilizing or maturation factors of NOX are presented in brown, activating binding partners are in green, complex organizing binding partners are in blue, and destabilizing binding partners are in red. In addition to the PM localization, NOX1 was also found in caveolae (71). NOX2 is heavily expressed in the plasma membrane of phagocytic vesicles. NOX4 was found in several sub-cellular compartments' membranes, for example, mitochondria (1, 18, 91). A soluble NOX4 splice variant, NOX4D (14, 62, 130), lacking five out of six transmembrane domains, was suggested in the nucleus and nucleolus (4). Although localization of NOX4 (135) and NOX5 (9, 170) in the ER was suggested, a physiological localization or associated function was not shown until now. Activation of NOX1–3 depends on the formation of hetero oligomeric complexes. NOX1 is activated by the binding of its organizer NOXO1, which along with the small GTPase, Rac, enables the binding of the NOXA1 to fully activate the complex. NOX2 is activated in a similar manner by its organizer proteins, p47phox and Rac, that enable binding of the activator protein, p67phox. The activation of NOX2 can be further enhanced by the binding of p40phox to the complex. Although NOX3 requires NOXO1 for activation (87), its requirement of activator proteins is still under debate but likely. NOXA1 seems to be capable of activating NOX3, but its role still needs to be confirmed in vivo (31, 32, 121, 171, 172). NOX4 is the only NOX isoform that seems to be constitutively active in the absence of any cytosolic binding factor. However, its activity can be enhanced by binding proteins such as protein Poldip2 (102) and activated TLR4 (14, 130, 168). TLR-4 also seems to bind to the NOX4D splice variant (14). Recently, an analogue of NOXO1, the Tks4/5, and PDI have been found to bind and activate NOX1 and NOX4 (43, 57). Hsp90 was shown to enhance the activities of NOX1, NOX2, and NOX5; while Hsp70 binds to NOX2 and NOX5, leading to degradation of the protein by ubiquitination (28, 29). However, the roles of the latter two binding proteins need further confirmation. NOX5, NOX6/DUOX1, and NOX7/DUOX2 are mainly activated by calcium via their calcium binding sites. The calcium sensitivity of NOX5 can be enhanced by calmodulin (170) and Hsp90 (28). Up to date, no calcium sensitizing or other binding partners of DUOX1/NOX6 or DUOX2/NOX7 have been identified. DUOX, dual oxidase; ER, endoplasmic reticulum; Hsp, heat shock protein; NADPH, nicotinamide adenine dinucleotide phosphate; NOX, catalytic subunit of NADPH oxidases; NOXA1, NOX activator-1; NOXO1, NOX organizer-1; PDI, protein disulfide isomerase; PM, plasma membrane; Poldip2, polymerase (DNA-directed) delta-interacting protein 2; TKS4/5, tyrosine kinase substrate with 4/5 SH3 domains; TLR4, toll-like receptor-4. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars