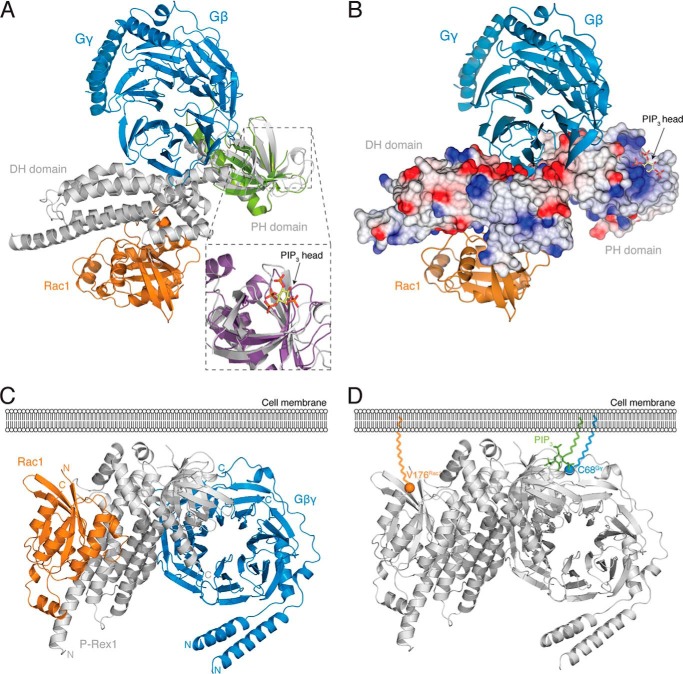
FIGURE 6.
Structural insights into P-Rex1·Gβγ and P-Rex1·PIP3 binding. A, alignment of the PH domain of P-Rex1 with the GRK2 PH·Gβγ complex (green; PDB code 1OMW (34), with an r.m.s. deviation of 2.046 Å across 93 residues) positions the Gβγ heterodimer on the opposite side of P-Rex1 from Rac1. In the alignment, Gβγ contacts both the P-Rex1 DH and PH domains. Inset, shows PIP3 modeled by structural alignment of the P-Rex1 PH domain with the CENTA1 PH·PIP3 complex (purple; PDB code 3LJU (65), with an r.m.s. deviation of 1.815 Å over 86 residues). B, electrostatic surface of P-Rex1 in the same orientation as shown in A, highlighting a highly negatively charged surface patch on P-Rex1 at the site of Gβγ binding and a highly positively charged pocket, where modeling places the negatively charged PIP3 head group. C and D, the membrane-interacting regions of each component of the P-Rex1·Rac1·Gβγ tetramer are located on the same face of the complex (C), including the C terminus of Rac1 extending from Val-176 (orange), the PIP3-binding pocket of the P-Rex1 PH domain, and the C-terminal Cys-68 of Gγ (blue) (D). PIP3 (green) shown as modeled in A. Val-176 of Rac1 and Cys-68 of Gγ are highlighted as spheres.