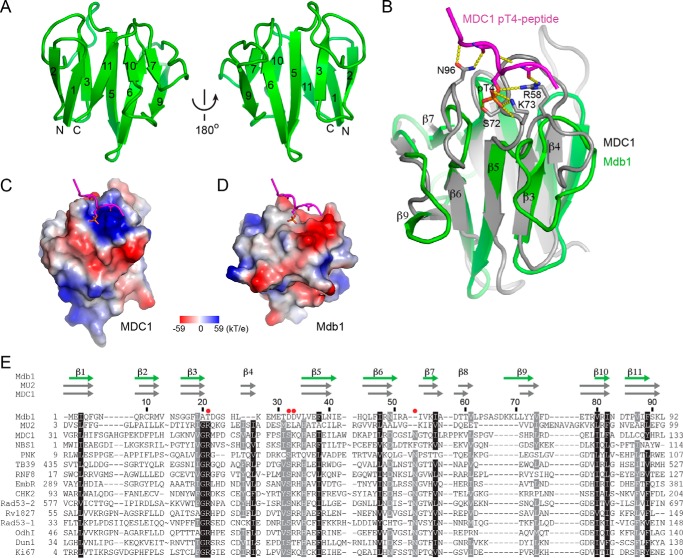
FIGURE 2.
Structure of the FHA domain of Mdb1. A, ribbon representation of the FHA domain of Mdb1. Two opposite views are displayed. The β-strands are numbered according to structural equivalence to those of canonical FHA domains. The N and C termini are labeled. B, Mdb1-FHA lacks a Thr(P)-binding pocket. The structure of Mdb1-FHA is superimposed with the structure of MDC1-FHA bound with the MDC1-Thr(P)-4 peptide (Protein Data Bank code 3UNN). The interacting residues in the MDC1 FHA-phosphopeptide complex are shown in a stick and ball representation with phosphorus colored yellow, oxygen red, and nitrogen blue. Hydrogen bonds are shown as dashed lines. C and D, charge surfaces of the FHA domains of MDC1 (C) and Mdb1 (D). The surfaces are colored from blue to red for positively to negatively charged regions. The two structures have the same orientation as in B. The phosphopeptide ligand of MDC1 is displayed in the Mdb1 structure to indicate the region corresponding to the Thr(P)-binding pocket in MDC1. E, structure-based sequence alignment of FHA domains. Aligned are FHA domains from the following (Protein Data Bank codes are shown in parentheses): Mdb1 (4S3H), MU2 (3UV0), MDC1 (3UNN), NBS1 (3HUF), PNK (2W3O), TB39.8 (3POA), RNF8 (2PIE), EmbR (2FF4), CHK2 (1GXC), Rad53-FHA1 (1G6G), Rv1827 (2KFU), Rad53-FHA2 (1J4L), OdhI (2KB3), Dun1 (2JQL), and Ki67 (2AFF). Except for Mdb1 and MU2, all FHA domains bind a phosphopeptide in structure. The key phosphopeptide-binding residues are marked with red solid cycles. The secondary structures are indicated for FHA domains of Mdb1 (green arrows), MU2 (gray arrows), and MDC1 (gray arrows) above the sequences. Residue numbers are shown for Mdb1. Omitted residues are indicated by ∼. Residues that are conserved in at least 90 and 80% of these sequences are shaded in black and gray, respectively.