FIGURE 4.
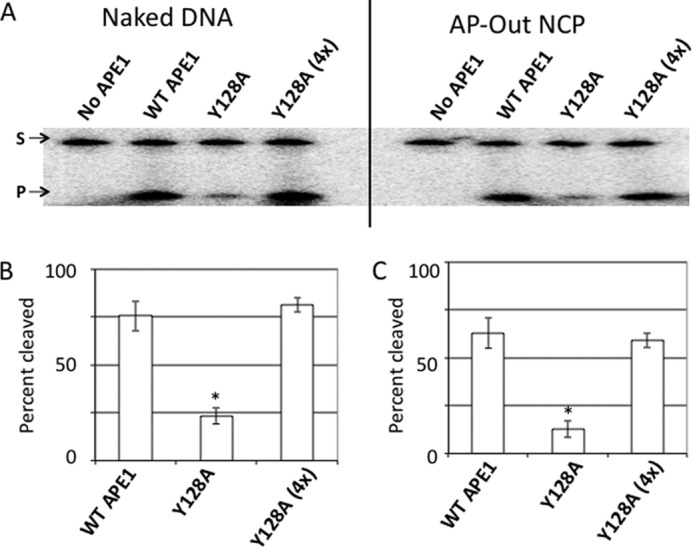
Activity of binding-defective APE1 mutant Y128A on nucleosome substrates. A, denaturing polyacrylamide gels separating the full-length 147-bp substrate (S, upper band) from the APE1 cleavage product (P, lower band) for the naked DNA (first four lanes), and the AP-Out nucleosome substrates (last four lanes). On naked DNA, the APE1 mutant Y128A was used at concentrations of 5 nm (as done for WT APE1), and at 20 nm (as Y128A (4x)). On AP-Out NCP substrates, WT APE1 and Y128A concentrations were 50 nm, and the Y128A (4x) was performed at 200 nm. B and C, charts showing percent cleavage of substrates (naked or AP-Out NCP DNAs) after a 10-min incubation with WT APE1 and Y128A at 5 and 20 nm (4x). Bars represent the average of at least 3 independent measurements, error bars represent standard deviations. Asterisks mark significant difference (as determined by t test) between the measurement of the variant and the corresponding measurement of the WT APE1, with p values as follows: Y128A naked DNA, p < 2 × 10−4; Y128A AP-Out Nucleosomes, p < 2 × 10−5.
