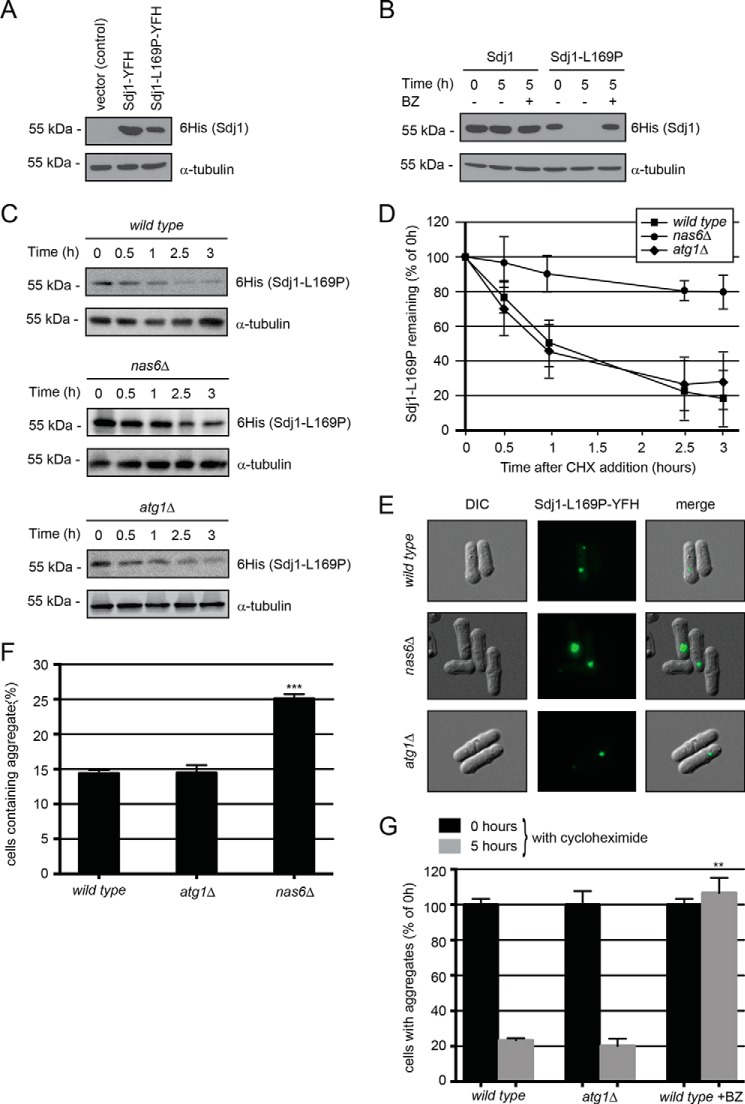
FIGURE 3.
Sdj1-L169P is degraded by the ubiquitin-proteasome system. A, the steady-state levels of Sdj1-YFH, Sdj1-C111A-YFH, and Sdj1-L169P-YFH were compared by SDS-PAGE and Western blotting using antibodies to His6 (to detect the YFH tag on the Sdj1 variants) and as a loading control α-tubulin. B, the amount of Sdj1-YFH or Sdj1-L169P-YFH was followed in cultures treated with cycloheximide (CHX) for 5 h. To some cultures, 1 mm of the proteasome inhibitor bortezomib (BZ) was also added. Equal loading was checked using antibodies to tubulin. C, the amount of Sdj1-L169P-YFH was followed in wild type, nas6Δ, and atg1Δ strains treated with cycloheximide. Equal loading was checked using antibodies to tubulin. D, quantification of degradation experiments as in C. Wild type, filled square; nas6Δ, filled circle; atg1Δ, filled diamond. The error bars indicate the S.E. ± mean (n = 4). E, fluorescence micrographs showing the Sdj1-L169P containing aggregates in the indicated genetic backgrounds. Note the larger aggregates in the nas6Δ strain. F, the number of the Sdj1-L169P-YFH aggregate containing cells was determined by fluorescence microscopy for the indicated strains. The error bars indicate the S.E. ± mean (n = 3, ***, p < 0.001, Student's t test). G, the stability of Sdj1-L169P aggregates in the indicated strains was determined by counting the number of cells containing aggregates in cultures treated with cycloheximide. The number of aggregate containing cells at 0 h was normalized to 100%. The error bars indicate the S.E. ± mean (S.E.) (n = 3, ***, p < 0.001, Student's t test). Before normalization the data were: wild type (0 h), 14.38 ± 0.47 (S.E.); wild type (5 h), 3.35 ± 0.18; atg1Δ (0 h), 14.45 ± 1.11; atg1Δ (5 h), 2.91 ± 0.60; wild type + BZ (0 h), 14.38 ± 0.47; wild type + BZ (5 h), 15.32 ± 1.24.