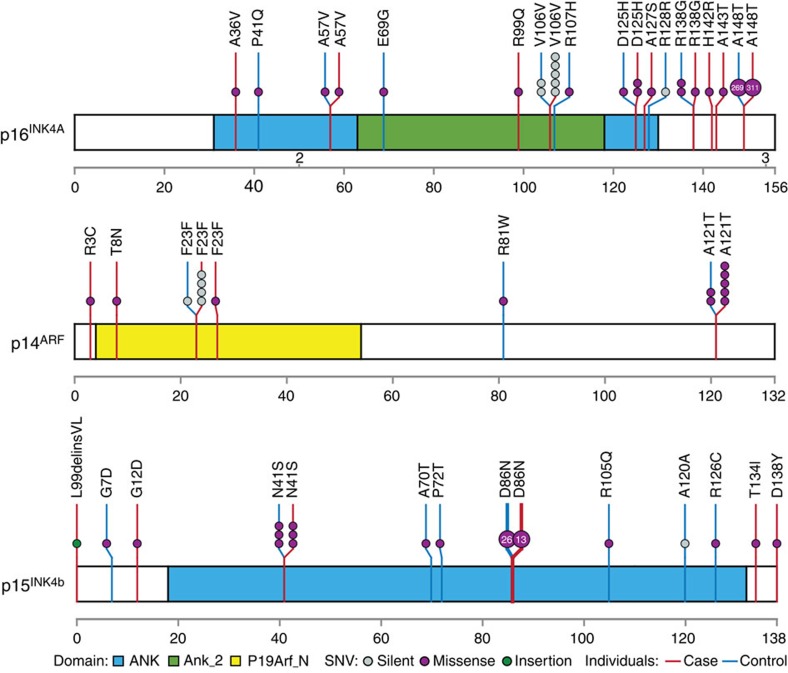
Figure 3. Targeted resequencing of CDKN2A–CDKN2B locus identified additional germline coding variants in children with ALL.
CDKN2A and CDKN2B genes were sequenced using Illumina HiSeq platform following capture-based enrichment of this genomic region in 2,407 ALL cases of European descent. Variants in non-ALL controls were based on publicly available data from the individuals of European descent within the NHLBI Exome Sequencing Project (N=4,300). Exonic variants are classified as silent or missense (grey or purple solid circles) and are mapped to three distinct open reading frames at this locus: p16INK4A, p14ARF and p15INK4B, for ALL cases (red vertical lines) and non-ALL controls (blue vertical lines), and functional domains are indicated by colour based on Pfam annotation. Each circle represents a unique individual carrying the indicated variant (heterozygous or homozygous), except for variants recurring in more than 10 individuals for which the number in the circle indicates the exact frequency of the observed variant.