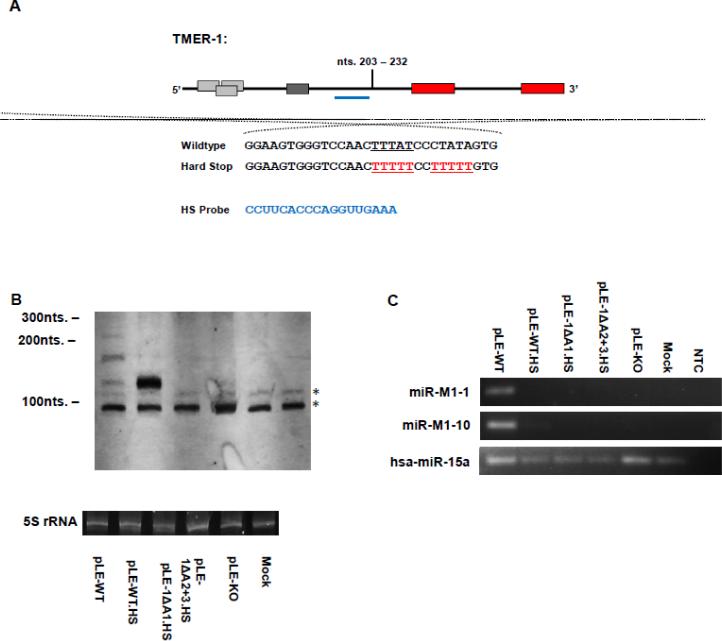
Figure 5. The impact of canonical RNA pol III termination signals near the predicted tRNA domain of TMER-1 on transcription and miRNA production from the TMER-1 gene.
(A) Schematic of TMER-1. Shown is the location and sequence of the hard stop mutation. The alternative transcriptional stop location in wildtype TMER-1 is underlined in the wildtype sequence. The hard stop sequences and locations generated in the hard stop mutant plasmids are underlined and in red below the wildtype sequence. The location of the hard stop probe use for northern analysis of TMER-1 in this figure is shown as a blue line below the TMER-1 diagram. The sequence of the TMER-1 hard stop probe in shown in blue from 3’ to 5’. (B) Northern blot for TMER-1 using a probe immediately downstream from the tRNA region of the TMER-1 transcript. Non-specific bands are indicated with asterisks. Below each blot is the ethidum bromide stained 5S rRNA band from each gel photographed to gauge RNA quality and loading prior to transfer. Labeled below each gel is the transfection condition from which the total RNA was isolated. (C) RLM-RT-PCR for viral miRNAs associated with the transcription of γHV68 TMER-1. The human miRNA, hsa-miR-15a is included as a cellular control for the RLM-RT-PCR assay. NTC – non-template control.