Abstract
The scavenger decapping enzyme hydrolyses the protecting 5′ cap structure from short mRNAs that result from exosomal degradation. Based on static crystal structures and NMR data it is apparent that the dimeric enzyme has to undergo large structural changes to bind substrate in a catalytically competent conformation. Here, we study the yeast enzyme and show that the associated opening-closing motions can be orders of magnitude faster than the catalytic turnover rate. This excess of motion is induced by binding of a second ligand to the enzyme, which occurs under high substrate concentrations. We designed a mutant that disrupts the allosteric pathway that links the second binding event to the dynamics and show that this mutant enzyme is hyperactive. Our data reveals a unique mechanism of substrate inhibition, where motions that are required for catalytic activity also inhibit efficient turnover, when they are present in excess.
Introduction
Enzyme catalysis is a fine-tuned balance between protein structure and dynamics 1-3. Over the past decades, our knowledge regarding protein structure has grown significantly 4. At the same time, our understanding of how fluctuations of the protein structure modulate function lags far behind 5-8. While indirect information about structural changes during catalysis can be gained from crystal structures of enzymes at different states along the reaction pathway, these static structures do not provide information about the rates with which an enzyme samples these different states. These rates have important implications for catalytic activity, however, only in very few cases it has been possible to directly link protein dynamics with function 9-14. Addressing this relationship experimentally is pivotal to advance our understanding of enzyme function. Importantly, conformational fluctuations also have implications for enzyme design 15 and for the development of pharmaceutical compounds that modulate catalytic activity.
In eukaryotes, mRNA is protected against degradation by the presence of a 3′ poly(A) tail and a 5′ cap structure (Supplementary Results, Supplementary Fig. 1a). The scavenger decapping enzyme (DcpS in humans; Dcs1p in yeast) catalyses cap hydrolysis of short (< 10 nucleotides) mRNAs that are produced by 3′ to 5′ exosomal mRNA degradation 16,17. The products of the reaction are m7G monophosphate (m7GMP) and an RNA body that carries a 5′ diphosphate (Supplementary Fig. 1a). The activity of the DcpS enzyme must be regulated to prevent premature decapping and unintentional degradation of an mRNA transcript. In yeast the Dcs2p protein can modulate the catalytic activity of the Dcs1p enzyme by direct interactions 18. Recently, the scavenger decapping enzyme gained interest as a target for the treatment of spinal muscular atrophy (SMA) 19,20.
DcpS is an 80 kDa homo-dimeric enzyme for which the human complex has been crystallized in its ligand free form and in the presence of minimal cap substrates (e.g. m7GpppG) (Supplementary Fig. 1) as well as in complex with pharmaceutically relevant inhibitors 19,21-23. DcpS is composed of a large dimeric C-terminal domain (50 kDa) that is flexibly connected by a hinge region to a smaller domain swapped dimeric N-terminal lid-like domain (30 kDa) (Supplementary Fig. 1b). In the substrate free form, the enzyme has a symmetric conformation with few contacts between the two domains 21. However, upon interaction with substrate, the complex adopts an asymmetric conformation, where one substrate is sandwiched between the N- and C-terminal domains on one side of the dimer (referred to as the closed site) 21,22. In this binding site, the catalytic triad is assembled to promote hydrolysis of the substrate 17,24. The other side of the dimeric enzyme adopts an open and catalytically incompetent conformation that can interact with a second substrate (referred to as the open site).
These static structures 21,22 together with biochemical experiments 25 and molecular dynamics simulations 26, have led to the view that DcpS undergoes large domain rearrangements during the catalytic cycle, both to interact with substrate and to release products. Presumably, the N-terminal lid-domain flips over during catalysis. This then results in an opening of the closed site and a simultaneous closing of the open site 22. This mechanism suggests that the dimeric nature of the enzyme and its domain motions enhances catalytic efficiency, as substrate can be positioned in the open binding site while catalysis is taking place at the closed binding site. However, several lines of evidence are in conflict with this model. First, single point-mutations that enhance activity have been identified, showing that the wild-type (WT) enzyme does not function at the maximally possible catalytic rate 21,22. Secondly, the DcpS enzyme is substrate inhibited, as it is more active under low substrate concentration than under high substrate concentration 25. This phenomenon is suggested to be due to a change in the rate-limiting step of the reaction from substrate binding (under single turnover conditions) to a conformational change in the enzyme (under multiple turnover conditions) 25. Quantitative data to test these hypotheses are not available and a more elaborate model that accurately explains the behaviour of the enzyme is required.
To gain insights into the catalytic cycle of the scavenger decapping enzyme and its regulation we here combined methyl group NMR spectroscopy, X-ray crystallography, and isothermal titration calorimetry (ITC) studies. This enabled us to reveal an unexpected view of how inter-domain motions directly regulate catalytic turnover and provides a unique example for the fact that regulatory mechanisms can be hidden in static structures. In brief, our results show that the domain flipping motions in the scavenger decapping enzyme are important for substrate binding and product release. We show that these motions increase with increasing substrate concentration and can reach frequencies that are two orders of magnitude faster than substrate turnover. Unexpectedly, these excessive flipping motions ultimately inhibit substrate hydrolysis. In agreement with that, we show that the hyperactivity of the K126A mutant enzyme is due to reduced motions of the lid domain. In addition, our data establish a unique mode of substrate inhibition in DcpS where high substrate concentrations induce excessive conformational changes in the enzyme that are too fast to permit catalysis.
Results
The free scavenger decapping enzyme is symmetric
To gain detailed information on the structure and dynamics of the scavenger decapping enzyme along the catalytic pathway, we first studied the ligand-free complex in solution by NMR spectroscopy. Methyl-TROSY NMR techniques 27 are ideally suited to probe motions in protein complexes and are applicable to large and complex systems 28-31. In this study, we used the scavenger decapping enzyme Dcs1p from S. cerevisiae, as the human enzyme for which X-ray structures are available did not provide the long-term stability required for detailed NMR studies. To prevent hydrolysis of the substrate during measurements we used a catalytically inactive variant of the enzyme (H268N)22. In our NMR experiments, we observed that the ligand-free form of the yeast DcpS enzyme displays only a single set of resonances (Supplementary Fig. 1d). This is consistent with both protein chains of the dimer being in an identical chemical environment, which is only the case in a symmetric homo-dimeric complex. Furthermore, we observed that the methyl-TROSY spectra of the isolated dimeric N- and C-terminal domains superimpose very well with the full-length protein (Supplementary Fig. 1d). This shows that there are no close contacts between the dimeric N-domain and the dimeric C-domain. In summary, we established that the substrate-free enzyme from yeast is symmetric in solution and that the N and C-terminal domains behave as independent units. These results are in full agreement with X-ray 21 and computational studies 26 of the ligand-free human enzyme that show that the angle between the N- and C-domains can change, but that the domains themselves move as rigid bodies.
DcpS interacts with substrates in a sequential manner
High-resolution structures are a prerequisite for the analysis of NMR studies that probe dynamics and interactions. The locally low sequence identity between the yeast and human DcpS enzymes precluded the generation of a reliable homology model for the yeast protein. We therefore used X-ray crystallography to determine the structure of the yeast DcpS enzyme (Dcs1p; H268N) in complex with m7GDP. We choose to use m7GDP as a ligand because it mimics the substrate and is not hydrolysed efficiently 32. We solved the structure of the yeast enzyme at a resolution of 2.3 Å (Table S1) and it features the same architecture as the structure of the human DcpS enzyme bound to m7GpppG as a substrate (1.8 Å rmsd over all Cα atoms in common secondary structure elements) (Fig. 1a, Supplementary Fig. 1c). As in the human protein, yeast DcpS adopts an asymmetric conformation where the ligand is placed between the N- and C-domains in the closed binding site, while the other side of the dimer adopts an open conformation (Fig. 1a, b). It is worth noting that the second site is occupied with substrate in the structure of the human enzyme 22 but is empty in our structure, which was crystallized at lower substrate concentration. This finding indicates that both binding sites in the asymmetric form of the enzyme differ significantly in affinity.
Figure 1. Structure of the yeast DcpS:substrate complex.
(a) 2.3 Å resolution crystal structure of the Dcs1p enzyme from S. cerevisiae in complex with m7GDP. The dimeric enzyme adopts an asymmetric conformation with one closed and one open binding site. The protein chains are coloured blue with magenta side chains and green with yellow side chains respectively and the substrate is coloured red. The boxed region is shown in panel b in a similar orientation. A cartoon representing the conformation of the enzyme is indicated, where the red dot refers to the substrate.
(b) The substrate is tightly bound in the closed binding site. Residues Y94 and W161 highlight the high asymmetry of the enzyme in the ligand bound form; these aromatic rings come within 5 Å of each other in the closed binding site, but are more than 22 Å apart in the open binding site. Without conformational changes, the substrate (product) is not able to dissociate from the enzyme.
To probe the binding mechanism between Dcs1p (H268N) and its substrate m7GpppG in solution, we used methyl-TROSY NMR spectroscopy to directly follow structural changes in the enzyme upon stepwise addition of the substrate (Fig. 2). In order to map the residues involved in this interaction, we assigned numerous isoleucine and methionine methyl resonances that are close to the active site or that can potentially probe conformational changes during the catalytic cycle 33,34 (See Supplementary Fig. 2). Interestingly, our NMR titration experiments display a two-step mechanism of substrate binding to DcpS: First, we observed that a large number of methyl resonances split into two signals upon addition of an equimolar amount of m7GpppG (e.g. I12, I36, I150, M153 and I219). This demonstrates that the initially symmetric enzyme converted into an asymmetric dimer upon complex formation with the substrate (Fig. 1, 2b). It is worth noting that the change in chemical environment between the symmetric and asymmetric conformation is much larger for the closed side than for the open side. We therefore attribute methyl resonances that display large chemical shift perturbations (CSPs) upon substrate addition to residues in the closed binding site, while those in the open side remained in many instances unaffected (Fig. 2b). The fact that the observed CSPs are in slow-exchange on the NMR time-scale strongly suggests that this first binding event occurs with high affinity. Upon addition of hyperstoichiometric concentrations of m7GpppG we observed additional CSPs for residues that are exclusively located in the open binding site of the asymmetric enzyme (Fig. 2c). Hence we can conclude that this second binding event takes place in the open side of the asymmetric dimer. Taken together, our NMR data are consistent with a sequential binding mechanism where the first substrate induces an asymmetric conformation of the enzyme and forms a closed binding site. Under higher substrate concentrations the second binding site on the open side of the asymmetric dimer can then be occupied, albeit with a lower affinity.
Figure 2. The scavenger decapping enzyme binds two substrates in a sequential manner.
(a) Methyl-TROSY NMR spectrum of the free, symmetric enzyme (0.25 mM dimer concentration). Assignments for a number of isoleucine and methionine residues that are close to the active site are indicated (see also Supplementary Fig. 2). Cartoons indicate the state of the complex.
(b) Methyl-TROSY NMR spectrum of Dcs1p after addition of one molar equivalent of m7GpppG (0.25 mM) to the dimeric enzyme. A number of resonances that report on the symmetric to asymmetric conversion are indicated with arrows.
(c) As in (b), but after addition of 20 molar equivalents of substrate (5 mM). Resonances that result from residues in the open binding site shift, indicating that a second substrate interacts in the open binding site of the enzyme. The insets on the right show a detailed view of the sequential binding process.
(d) Plot of the changes in the peak intensities for residue I12 (open and closed resonances) and the changes in the peak position for residue M153 (CSP in carbon) for 15 titrations points (see a-c and boxed regions in the spectra of panels b and c). The errors (s.d.) in the extracted binding constants are based on Monte Carlo simulations for a simultaneous fit of multiple residues (Supplementary Fig. 3a). For clarity the scale of the x-axis is shown different below and above the 1:1 m7GpppG:Dcs1p ratio, as is indicated by the waveform.
DcpS contains a high- and a low-affinity binding site
In order to quantify the affinities associated with these two binding events we performed accurate NMR titration experiments (Fig. 2d, Supplementary Fig. 3a). We then used intensities of the resonances that report on the first binding event (I12, I36, Fig. 2b) and the position of the resonances that report on the occupancy of the second binding site (I42, M153, Fig. 2c) to extract dissociation constants for both binding events. To that end we fitted the spectral properties using a sequential binding model (Supplementary Fig. 3a). From this, we extracted a high nano-molar affinity for the first binding event (68 ± 255 nM) and a three orders of magnitude weaker micro-molar affinity for the second binding event (105 ± 20 μM) (Table 1). A high-affinity binding event was previously reported in the human enzyme 25,35, whereas the low-affinity binding event had not been observed before. To independently validate the sequential binding and the large difference in affinity for the two binding events we turned to ITC measurements (Supplementary Fig. 3a). In the ITC thermograph a first binding event was observed in the nano-molar range, whereas the affinity for the second binding site was determined reliably (324 ± 67 μM), in agreement with our NMR data. Taken together, our NMR and ITC data demonstrate that the scavenger decapping enzyme interacts with two substrates in a sequential manner, where the second binding site emerges only after interaction of the enzyme with the first substrate.
Table 1. Summary of the measured affinities, domain flipping rates and turnover rates.
| WT (H268N) | K126A (H268N) | |
|---|---|---|
|
| ||
| Kd substrate (m7GpppG) | Closed site: 68 (255) nM a,c |
Closed site: 112 (390) nM a,c 240 (90) nM b |
|
Open site: 105 (20) μM a 324 (67) μM b |
Open site: 196 (31) μM a 280 (30) μM b |
|
|
| ||
| Flipping rate | Ranging from 8 Hz (low substrate excess) to 34 Hz (high substrate excess) |
< 1 Hz under high and low substrate excess |
|
| ||
| Turnover rate | 0.27 s−1 under high substrate excess |
0.48 s−1 under high substrate excess |
determined using NMR
determined using ITC
the high uncertainty in the determined kD of the closed binding site results from the limited number of data points that define the transition.
The C-terminal domain binds the second substrate
To characterise the second binding site in more detail, we performed ITC and NMR experiments with the isolated C-terminal domain of the yeast Dcs1p enzyme and m7GpppG. Using ITC, we showed that the isolated C-terminal domain of the enzyme interacts with m7GpppG with an affinity that is comparable to that of the second binding event in the full-length complex (Supplementary Fig. 3c). Together with the structure of the human DcpS enzyme 22 (Supplementary Fig. 1b, 3c) this shows that the second ligand interacts predominantly with the C-terminal domain in the open binding pocket. In order to structurally characterize the substrate binding to the open site of the yeast enzyme, we performed NMR titration experiments using the isolated dimeric C-terminal domain and compared these to the titration experiment performed with the full-length enzyme. For the isolated C-terminal domain, we observed that the same residues experience chemical shift perturbations upon addition of substrate as in the full-length protein at hyperstoichiometric substrate concentrations (Supplementary Fig. 3c). Importantly, despite the fact that the second ligand mainly contacts the C-terminal domain, we observed that occupation of the second binding site in the full-length protein results in CSPs in the N-terminal domain (e.g. I42; Fig. 2c). Based on the structure of the human protein in complex with m7GpppG substrate, the distance between the second substrate and I42 in the yeast enzyme is expected to be over 12 Å. This long range effect points towards an allosteric mechanism that can relay the substrate binding event in the open binding pocket to the N-terminal lid domain and potentially further towards the closed binding pocket.
Activity requires the asymmetric form of the complex
Both X-ray (Fig. 1) and NMR (Fig. 2) data strongly suggest that the scavenger decapping enzyme needs to adopt an asymmetric form in order to be catalytically competent. In yeast DcpS (Fig. 1), this closed conformation is mediated by a network of interactions that involve contacts between the 7-methyl group of the substrate and the aromatic ring of tyrosine 94. To assess the importance of these interactions for stable domain closure we recorded methyl NMR spectra of the Y94A (H268N) enzyme in presence of m7GpppG and of the WT (H268N) enzyme in presence of the unmethylated ligand GpppG (Supplementary Fig. 4a). In both cases, we observed no resonances corresponding to the asymmetric conformation (Supplementary Fig. 4a), like we did for the WT (H268N) enzyme with m7GpppG (Fig. 2b). However, we did observe small CSPs in the same region as we observed for substrate binding to the WT protein (Fig. 2 and Supplementary Fig. 4a). This shows that impairing the DcpS Y94: 7-methyl interaction results in a situation where the enzyme substrate complex is no longer able to form a stable asymmetric conformation.
To directly test the importance of the asymmetric conformation for catalytic activity, we monitored the hydrolysis of m7GpppG and GpppG by the WT yeast DcpS enzyme and the Y94A mutant, respectively, using a time series of one-dimensional 31P NMR spectra. The WT DcpS enzyme hydrolyses m7GpppG into m7GMP and GDP (see Supplementary Fig. 1a, S4b), however, we did not detect enzymatic turnover for the Y94A enzyme with m7GpppG or for the WT enzyme with GpppG (Supplementary Fig. 4b). In summary, we show that the formation of a sufficiently long lived closed active site is crucial for catalytic activity, which underscores the importance of conformational changes in the catalytic cycle of the scavenger decapping enzyme.
A conformational change results in product release
The reaction products m7GMP and GDP are formed in the closed pocket of the scavenger decapping enzyme after hydrolysis of the m7GpppG substrate. Using methyl NMR spectroscopy, we observed that binding of the product m7GMP to the enzyme preserves the asymmetric form of the complex (Supplementary Fig. 5a). During the next step in the catalytic cycle, the enzyme complex thus needs to open in order to release the product. To determine the affinity of the product for the closed binding site, we performed ITC measurements and found that the m7GMP product binds to yeast DcpS in a single-step process (Kd = 1.1 ± 0.6 μM) (Supplementary Fig. 5b). The product thus interacts a factor 4 more weakly with the closed binding site than the substrate does. Interestingly, using ITC and NMR, we were not able to detect an interaction of the m7GMP product with the second, open binding site. After hydrolysis of the substrate in the closed binding site and a flipping motion of the enzyme the product occupies the open binding site and m7GMP will be efficiently released from the enzyme. To test this hypothesis we used methyl-TROSY NMR spectroscopy, where we added m7GpppG substrate to the m7GMP:Dcs1p complex. This resulted in the efficient release of product and the formation of an m7GpppG:Dcs1p complex and shows that the enzyme can indeed spontaneously change from a product bound state into a substrate bound state upon addition of substrate (Supplementary Fig. 5c). In summary, our results indicate that the first and second binding sites can concomitantly open and close during the catalytic cycle.
Dcs1p undergoes spontaneous domain flipping motions
To observe if the scavenger decapping enzyme undergoes domain flipping motions under excess m7GpppG substrate concentrations, we performed longitudinal exchange experiments 29,33,36 on highly deuterated methyl labelled samples. In these experiments the conversion of an open binding site into a closed one and vice versa can be observed directly, as the exchange process results in cross peaks at resonance frequencies where one dimension corresponds to the open (closed) state and the other dimension results from the closed (open) state. These experiments are possible as NMR is unique in the fact that it can distinguish between the open and closed states within one enzyme complex (Fig. 2b,c). Here, we observed exchange peaks for all residues that display a different chemical shift in the open and closed states, indicating that the enzyme undergoes continuous domain-flipping motions (Fig. 3a). Measurements using a number of different exchange delays in these experiments allow for the extraction of the exchange rates and the associated populations. For all residues that displayed well resolved resonances for the auto and cross peaks we determined very similar exchange parameters, indicating that the process we detect results from a global conformational change as is associated with the N-terminal domain flipping on the C-terminal domain. In addition, the extracted populations of the open and closed states were, as expected, equal, as the number of open and closed sites in the enzyme is by definition 1:1. During the further analysis of the data, we thus used a global fit, where all residues were fitted to one exchange process and where pclosed and popen were fixed to 0.5. In the presence of a 20-fold excess of substrate over enzyme we extracted a domain flipping rate of 35.4 (± 4.8) s−1 (Fig. 3b). Importantly, the flipping motions that we detected are not a result of enzymatic turnover and subsequent product release since an inactive mutant (H268N) was used. Rather, the motions report on a situation where an occupied closed binding site turns into an occupied open binding site, and vice versa.
Figure 3. Quantification of domain flipping motions in the scavenger decapping enzyme.
(a) Longitudinal exchange experiment that directly reports on motions in the enzyme where the open binding site closes and where the closed binding site opens. The cyan and pink coloured resonances result from the closed and open sites, respectively. The arrows point to the resonances that appear due to the domain flipping motions. The spectrum shown here is recorded with a mixing time of 75 ms in the presence of a 20-fold excess of substrate over the dimeric enzyme.
(b) Quantification of the exchange process in the presence of a 20 fold excess of ligand. The circles indicate resonance intensities of the cross and auto peaks. The error bars represent uncertainties in the resonance intensities. The drawn lines are a best global fit to the data and yield an exchange rate of 35.4 (± 4.8) s−1. The error (s.d.) in the extracted parameters are based on 100 Monte Carlo simulations.
(c) The flipping motions (a, b) depend on the m7GpppG ligand excess. Higher excess of ligand results in a higher occupation of the open second binding site (Figure 2c, d) and in faster exchange rates. This correlation between exchange rates and binding site occupancy shows that the flipping motions are directly induced by the presence of the second ligand in the open binding site.
Second substrate binding induces domain flipping motions
The second (open) binding site in Dcs1p is saturated with substrate (Fig. 2d) under the conditions where we observed N-domain flipping motions. To probe if the substrate in the open binding site induces these motions, we measured the domain flipping rates at decreasing substrate concentrations. Interestingly, the domain flipping rates reduce with reduced occupation of the second binding site (Fig. 3c). These results display the presence of an allosteric pathway that links both binding sites in the enzyme. Such an allosteric pathway has previously been suggested based on biochemical experiments 25, but the molecular basis underlying the cross talk between the active sites has not been known. Here we demonstrate that substrate binding in the open binding site of the enzyme directly induces domain rearrangements, causing the closed binding site to open and the open binding site to close.
The catalytic rates are slower than the flipping rates
To assess if the substrate-induced flipping motions in the enzyme correlate with catalytic turnover we determined the activity of Dcs1p in excess substrate conditions. To that end, we continuously recorded phosphorus NMR spectra during enzymatic turnover. The initial spectra (t=0) display the phosphor resonances of m7GpppG and the final spectra those of the products m7GMP and GDP (Fig. 4a). From these data we extracted a turnover rate (kcat) of 0.27 s−1 (Fig. 4b). This rate agrees with the turnover rate under high substrate concentrations that was previously determined for the human enzyme complex 25. However, it is important to note that the turnover rates at high substrate excess are two orders of magnitude slower than the domain flipping motions of 35.4 s−1 that we have measured (Fig. 3).
Figure 4. Turnover rates of the enzyme are much slower than the flipping rates.
(a) 31P NMR spectra that report on the Dcs1p mediated degradation (50 nM enzyme) of the (0.5 mM) m7GpppG substrate (red, top) into m7GMP and GDP product (bottom, yellow). NMR spectra (18 minutes each) were recorded successively until completion of the reaction.
(b) Progression curves of the reaction. Crosses correspond to the mean concentration of the substrate and product signals and are derived from the peak intensities of all 31P signals. The drawn line corresponds to the best fit of the data, the extracted turnover rates are indicated.
(c) Longitudinal exchange experiment of Dcs1p K126A in the presence of a 20 fold excess of substrate and using a mixing time of 75 ms (where the exchange peaks have maximum intensity for the WT protein). Exchange peaks were not observed (arrows in the traces) demonstrating that the K126A enzyme does not undergo unproductive flipping motions.
A point mutation interferes with fast flipping motions
Above we determined that the N-terminal domain senses the presence of substrate in the open binding pocket (Fig. 2c). The structure of the enzyme (Fig. 1) features positively charged amino acids such as K126 in the hinge region. We reasoned that these residues could be part of the allosteric pathway that links the occupation of the open binding site to a conformational change. To assess this hypothesis we mutated K126 to an alanine and used longitudinal exchange experiments to probe if the domain flipping motions in this mutant enzyme are altered. Indeed, under substrate excess, the domain motions that we detected in the WT enzyme are no longer detectable in the mutant enzyme (Fig. 4c). Based on the detection limits of the longitudinal exchange experiment, the domain flipping motions are reduced from 35.4 s−1 in the WT protein to less than 1 s−1 in the K126A mutant. It is important to note that the K126A mutant behaves like WT protein regarding the formation of an asymmetric conformation and regarding the interaction with substrate (Supplementary Fig. 3c). Indeed, the affinity for the substrate did not change significantly by introducing the K126A mutation in the enzyme (Table 1). We thus found that a single point mutation in Dcs1p is able to uncouple the binding of substrate in the second (open) binding pocket from fast domain rearrangements.
Reduced domain flipping motions lead to hyperactivity
To probe if reduced conformational exchange influences enzymatic activity we measured the activity of the K126A enzyme. Interestingly, we found that the K126A enzyme displays a two-fold higher activity (Fig. 4b) compared to the WT enzyme. It is important to note that the increased catalytic efficiency is not due to changes in substrate binding (Supplementary Fig. 3a, b and Table 1). In addition, potential changes in product release are not the cause of the observed hyperactivity, as for the human enzyme it is established that product release is not the rate-limiting step in the catalytic cycle 25.
Hyperactive mutants of DcpS are described for the human enzyme 22, however, the molecular basis for this hyperactivity remained elusive. Here, we show that unproductive domain flipping motions decrease the catalytic efficiency of the WT enzyme. Interference with the underlying allosteric pathway through mutations reduces the unproductive motions and results in hyperactivity. Hence, domain flipping motions in the enzyme that are important for the catalytic cycle (Supplementary Fig. 4, 5) can reduce activity when they are too fast (Fig. 3, 4).
Discussion
The correlation between motions in enzymes and catalytic activity remains poorly understood, despite the fact that this relationship has been proposed decades ago 37.
Here, we study dynamic changes during the catalytic cycle of the yeast scavenger decapping enzyme. Static structures of the enzyme 21,22 (Fig. 1, Supplementary Fig. 1) suggest that domain rearrangements are important for substrate binding and product release. We showed that the formation of a stable asymmetric conformation is indispensable for activity (Fig. 2, Supplementary Fig. 4, 5). In addition, we established that the interaction of substrate with the second (open) binding pocket of the dimeric enzyme induces unproductive motions that impair catalytic efficiency (Fig. 3, 4).
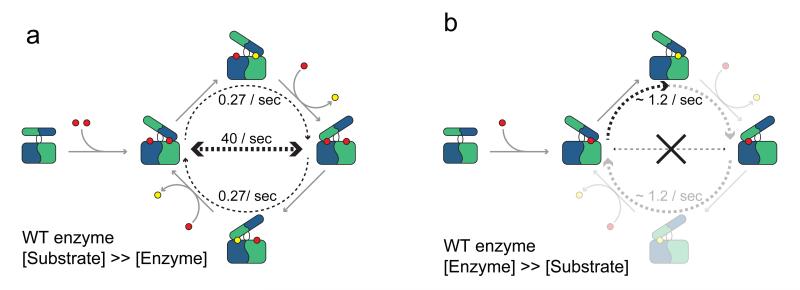
At high substrate concentrations the hinge motions interfere with efficient catalysis (Fig. 5a) as the time the substrate spends in the closed active site is on average too short to allow for efficient hydrolysis: before catalysis can take place the enzyme rearranges into its mirror form (Fig. 5). This implies that the rate-limiting step in the catalytic cycle is after the formation of the closed binding site, e.g. structural rearrangements in the substrate. Previous studies indicate that, for the human enzyme, such a conformational change is relevant for the activation of cap hydrolysis 22.
Figure 5. Cartoon representation of the substrate inhibition mechanism of the scavenger decapping enzyme.
(a) Under substrate excess, both binding sites in the enzyme are occupied. This results in fast non-productive flipping motions (horizontal dashed arrow) and low turnover rates (curved dashed arrows). Substrate and product is indicated with red and yellow circles respectively. Determined rates are indicated.
(b) Under single turnover conditions, only one of the two binding sites in the enzyme is occupied. The unproductive motions (crossed out horizontal line) no longer take place and the catalytic turnover is increased (curved dashed arrow). The indicated turnover rate corresponds to the human enzyme 25 and may differ for the yeast complex. Note that under these conditions part of the catalytic cycle is not sampled and indicated in a light colour for reference only.
Using a designed mutant enzyme (K126A), we uncoupled the second substrate binding event from the induction of unproductive motions, which resulted in hyperactivity (Fig. 4, Supplementary Fig. 6). Mechanistically, we propose that electrostatic interactions between the positively charged hinge region (K126 and in addition K125) and the negatively charged phosphate groups of the substrate in the open binding site induce motions of the N-domain. The conservation of positively charged residues in the hinge region points towards a preserved electrostatic mechanism that regulates the activity of the scavenger decapping enzyme family. In agreement with that, mutations of positively charged hinge residues in the human enzyme (e.g. K138) also leads to hyperactivity 22.
Previously, it was shown that the human DcpS enzyme is more active at low substrate concentrations than it is at high substrate concentrations 25. The physiological relevance of substrate inhibition has been demonstrated for a variety of enzymes, including phosphofructokinase, tyrosine hydroxylase and acetylcholinesterase 38. DcpS is part of a complex enzymatic network that eliminates mRNA cap structures 39 and substrate inhibition could play a role in a regulatory feedback loop. In addition, DcpS substrate inhibition might be required to limit cellular levels of the product m7GMP that can be misincorporated into RNA 18. The model of the catalytic cycle of DcpS that we introduced here (Fig. 5) provides the molecular explanation for the observed substrate inhibition (Fig. 5b). At low substrate concentrations the second DcpS binding site is not occupied (Fig. 2) and the domain flipping motions are significantly reduced or absent (Fig. 3, 5b). This results in an increase in the catalytic turnover rate that can reach 1.2 s−1 in the human enzyme 25. To our knowledge, this is the first time that a mechanism of substrate inhibition through excess of protein motions has been described.
The substrate induced molecular motions provide an additional layer of regulation for the activity of the scavenger decapping enzyme. In that regard it is worth mentioning that the enzyme has been reported to interact with a number of partners, including the exosome complex in humans 16 and Xrn1 40 and Dcs2p 18 in S. cerevisiae. Dcs2p has been reported to inhibit Dcs1p activity by heterodimerization, which potentially influences the dynamics in the complex. Currently, efforts to pharmaceutically inhibit DcpS are underway 19,20. We here demonstrate that DcpS operates in a narrow range of dynamics. Thus both small compounds that block inter-domain motions as well as those that accelerate these can impair activity. Finally, DcpS displays different activities towards substrates with various RNA lengths. The mechanism by which the enzyme discriminates short substrates from longer ones 35 might be through modulation of domain motions by the RNA body. Our results provide the conceptual framework to address these questions in detail.
In summary, we here present an example of an enzyme where conformational changes that are required for the activity can be detrimental for catalysis in case they are too fast. This surprising insight could not have been obtained based on the knowledge of static structures alone and underscores the need for studies that localize and quantify motions in large bio-molecular assemblies. We expect these studies to elucidate more instances of complex interrelationships between dynamics and activity, thus greatly advancing our understanding of bio-molecular function.
Online methods
Protein preparation
The Dcs1p gene from S. cerevisiae was amplified from genomic DNA and cloned into a modified pEt-vector that contained an N-terminal His6-NusA-His6 tag that could be removed using TEV protease. Mutations were introduced using standard quick-change methods. BL21 (DE3) codon plus cells were transformed with the respective plasmid and grown in rich or minimal medium in the presence of kanamycin and chloramphenicol. Minimal medium was based on 100% D2O and contained 2H12C glucose as a carbon source. Isoleucine and methionine labelling was achieved by addition of 50 mg L−1 α-ketobutyric acid and 250 mg L−1 of [methyl-13CH3]-methionine, respectively, one hour prior to induction with 1.0 mM IPTG at a cell density (OD600) of 0.8 and a temperature of 25°C. 12 hours after induction the cells were lysed in buffer A (50 mM NaPO4 pH 8.0, 150 mM NaCl and 5mM imidazole) supplemented with 0.1% Triton, 1mM EDTA and lysozyme. Subsequently the cell lysate was centrifuged at 50,000 g for 20 minutes to remove insoluble debris. The supernatant supplemented with 2 mM MgCl2 was applied to Ni-NTA resin and the column was washed using 10 column volumes of buffer A. The protein was eluted from the resin using buffer A supplemented with 200 mM imidazole. The affinity and solubility tag was removed from the protein by TEV cleavage during dialysis into 25 mM Tris pH 8.0, 75 mM NaCl, 1mM DTT. The cleaved tag was removed from the target protein using a second Ni-NTA step in dialysis buffer. Finally a size exclusion chromatography purification step in buffer B (25mM Hepes pH 8.0, 25 mM NaCl) using a Superdex 200 column yielded pure protein. For NMR the final protein samples were exchanged by successive concentration/ dilution steps into buffer B that was based on 100% D2O (NMR buffer).
Crystallography
Dcs1p in complex with m7GDP crystallized at a concentration of 10 mg/ml in 100 mM Hepes pH 7.5, 100 mM NaCl, 1.6 M AmSO4 after several weeks. Diffraction data were collected at 100 K using a wavelength of 1 Å and a PILATUS 6 M detector at beamline PXII of the Swiss Light Source (PSI, Villigen, Switzerland). Data were processed using XDS 41 and molecular replacement was performed using Phaser 42 with the crystal structure of the human protein in complex with m7GDP (1XMM)21 as a search model. The structure was finalized by iterative manual modeling with Coot 43 and refinement with Phenix 44 and Refmac 45. The crystalized protein contained a cysteine bridge in a loop region of the N-domain (Cys 72-Cys 78). This crystallization artifact does not interfere with the data analysis, as the activity of the enzyme does not change between oxidizing and reducing conditions (Supplementary Fig. 7). All figures displaying protein structures were generated using PyMOL (http://www.pymol.org/).
NMR
NMR spectra were recorded at 27°C on Bruker AVIII-600 and AVIII-800 spectrometers with room temperature probe-heads. All NMR samples contained buffer based on 100% D2O. For degradation experiments 0.5 mM m7GpppG substrate were mixed with 50 nM Dcs1p in NMR buffer and successive phosphorus experiments were recorded over a time of several hours. Proton-carbon based experiments exploited the methyl-TROSY principle and were recorded using a carbon chemical shift evolution of typically 40 ms. Longitudinal exchange experiments were recorded on samples containing 0.25 mM Dcs1p (dimer concentration) in the presence of a 1:1, 1:2.5, 1:5 and 1:20 molar ratio of m7GpppG using mixing times ranging from 3 to 1000 ms, depending on the exchange rate. NMR data were processed using the nmrPipe/nmrDraw software suite 46 and figures displaying NMR spectra were produced using nmrView (onemoonscientific.com). NMR binding affinities were determined as described in the legend to Supplementary Fig. 3a. All data points of the titration were simultaneously fitted to one sequential binding event. The extracted kD values indicate that the first binding event takes place in the high nano-molar range and that the second binding event has a dissociation constant of 105 (±20) μM. The error bars represent uncertainties in the peak intensity or peak position. The extracted kD values represent mean values ± s.d and are based on Monte Carlo simulations where the data points were randomly varied according to their errors. The large uncertainty in the kD value for the first binding event results from the limited number of titration points that define the event.
ITC
Binding constants were determined using a TA instruments nanoITC at 25°C. m7GpppG or m7GMP were injected into a 0.5 mM Dcs1p (all variants carried the H268N mutation) solution (either full length or C-domain). Data were processed using the in-house written scripts (see legend to Supplementary Fig. 3a). Errors in the extracted Kd values are based on Monte Carlo simulations where the individual heat emissions were randomly varied based on the noise level in the experiment.
Supplementary Material
Acknowledgments
We acknowledge Janina Peters for excellent technical support, Silke Wiesner for discussions, Georg Zocher for assistance in recording diffraction data, Thilo Stehle for support and Lewis Kay for valuable comments on the manuscript. This work has received funding from the Max Planck Society and the European Research Council under the European Union’s Seventh Framework Programme (FP7/2007–2013), ERC grant agreement no. 616052.
Footnotes
Competing financial interests
The authors declare no competing financial interests.
Additional information
Supplementary information is available in the online version of the paper.
Accession codes
Protein Data Bank: the coordinates for Dcs1p in complex with m7GDP are deposited under accession code 5BV3.
References
- 1.Fersht AR. Structure and mechanism in protein science: A guide to enzyme catalysis and protein folding. W. H. Freeman; 1999. [Google Scholar]
- 2.Hammes-Schiffer S, Benkovic SJ. Relating protein motion to catalysis. Annu Rev Biochem. 2006;75:519–41. doi: 10.1146/annurev.biochem.75.103004.142800. [DOI] [PubMed] [Google Scholar]
- 3.Boehr DD, Dyson HJ, Wright PE. An NMR perspective on enzyme dynamics. Chem Rev. 2006;106:3055–79. doi: 10.1021/cr050312q. [DOI] [PubMed] [Google Scholar]
- 4.Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henzler-Wildman K, Kern D. Dynamic personalities of proteins. Nature. 2007;450:964–72. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 6.Tokuriki N, Tawfik DS. Protein dynamism and evolvability. Science. 2009;324:203–7. doi: 10.1126/science.1169375. [DOI] [PubMed] [Google Scholar]
- 7.McGeagh JD, Ranaghan KE, Mulholland AJ. Protein dynamics and enzyme catalysis: insights from simulations. Biochim Biophys Acta. 2011;1814:1077–92. doi: 10.1016/j.bbapap.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 8.van den Bedem H, Fraser JS. Integrative, dynamic structural biology at atomic resolution-it’s about time. Nat Methods. 2015;12:307–18. doi: 10.1038/nmeth.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eisenmesser EZ, Bosco DA, Akke M, Kern D. Enzyme dynamics during catalysis. Science. 2002;295:1520–3. doi: 10.1126/science.1066176. [DOI] [PubMed] [Google Scholar]
- 10.Boehr DD, McElheny D, Dyson HJ, Wright PE. The dynamic energy landscape of dihydrofolate reductase catalysis. Science. 2006;313:1638–42. doi: 10.1126/science.1130258. [DOI] [PubMed] [Google Scholar]
- 11.Henzler-Wildman KA, et al. Intrinsic motions along an enzymatic reaction trajectory. Nature. 2007;450:838–44. doi: 10.1038/nature06410. [DOI] [PubMed] [Google Scholar]
- 12.Wolf-Watz M, et al. Linkage between dynamics and catalysis in a thermophilic-mesophilic enzyme pair. Nat Struct Mol Biol. 2004;11:945–9. doi: 10.1038/nsmb821. [DOI] [PubMed] [Google Scholar]
- 13.Zavodszky P, Kardos J, Svingor, Petsko GA. Adjustment of conformational flexibility is a key event in the thermal adaptation of proteins. Proc Natl Acad Sci U S A. 1998;95:7406–11. doi: 10.1073/pnas.95.13.7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agarwal PK, Billeter SR, Rajagopalan PT, Benkovic SJ, Hammes-Schiffer S. Network of coupled promoting motions in enzyme catalysis. Proc Natl Acad Sci U S A. 2002;99:2794–9. doi: 10.1073/pnas.052005999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rothlisberger D, et al. Kemp elimination catalysts by computational enzyme design. Nature. 2008;453:190–5. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- 16.Wang Z, Kiledjian M. Functional link between the mammalian exosome and mRNA decapping. Cell. 2001;107:751–62. doi: 10.1016/s0092-8674(01)00592-x. [DOI] [PubMed] [Google Scholar]
- 17.Liu H, Rodgers ND, Jiao X, Kiledjian M. The scavenger mRNA decapping enzyme DcpS is a member of the HIT family of pyrophosphatases. EMBO J. 2002;21:4699–708. doi: 10.1093/emboj/cdf448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malys N, McCarthy JE. Dcs2, a novel stress-induced modulator of m7GpppX pyrophosphatase activity that locates to P bodies. J Mol Biol. 2006;363:370–82. doi: 10.1016/j.jmb.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 19.Singh J, et al. DcpS as a therapeutic target for spinal muscular atrophy. ACS Chem Biol. 2008;3:711–22. doi: 10.1021/cb800120t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gogliotti RG, et al. The DcpS inhibitor RG3039 improves survival, function and motor unit pathologies in two SMA mouse models. Hum Mol Genet. 2013;22:4084–101. doi: 10.1093/hmg/ddt258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen N, Walsh MA, Liu Y, Parker R, Song H. Crystal structures of human DcpS in ligand-free and m7GDP-bound forms suggest a dynamic mechanism for scavenger mRNA decapping. J Mol Biol. 2005;347:707–18. doi: 10.1016/j.jmb.2005.01.062. [DOI] [PubMed] [Google Scholar]
- 22.Gu M, et al. Insights into the structure, mechanism, and regulation of scavenger mRNA decapping activity. Mol Cell. 2004;14:67–80. doi: 10.1016/s1097-2765(04)00180-7. [DOI] [PubMed] [Google Scholar]
- 23.Han GW, et al. Crystal structure of an Apo mRNA decapping enzyme (DcpS) from Mouse at 1.83 A resolution. Proteins. 2005;60:797–802. doi: 10.1002/prot.20467. [DOI] [PubMed] [Google Scholar]
- 24.Seraphin B. The HIT protein family: a new family of proteins present in prokaryotes, yeast and mammals. DNA Seq. 1992;3:177–9. doi: 10.3109/10425179209034013. [DOI] [PubMed] [Google Scholar]
- 25.Liu SW, Rajagopal V, Patel SS, Kiledjian M. Mechanistic and kinetic analysis of the DcpS scavenger decapping enzyme. J Biol Chem. 2008;283:16427–36. doi: 10.1074/jbc.M800341200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pentikainen U, Pentikainen OT, Mulholland AJ. Cooperative symmetric to asymmetric conformational transition of the apo-form of scavenger decapping enzyme revealed by simulations. Proteins. 2008;70:498–508. doi: 10.1002/prot.21540. [DOI] [PubMed] [Google Scholar]
- 27.Tugarinov V, Hwang PM, Ollerenshaw JE, Kay LE. Cross-correlated relaxation enhanced 1H[bond]13C NMR spectroscopy of methyl groups in very high molecular weight proteins and protein complexes. J Am Chem Soc. 2003;125:10420–8. doi: 10.1021/ja030153x. [DOI] [PubMed] [Google Scholar]
- 28.Sprangers R, Kay LE. Quantitative dynamics and binding studies of the 20S proteasome by NMR. Nature. 2007;445:618–22. doi: 10.1038/nature05512. [DOI] [PubMed] [Google Scholar]
- 29.Audin MJ, et al. The archaeal exosome: identification and quantification of site-specific motions that correlate with cap and RNA binding. Angew Chem Int Ed Engl. 2013;52:8312–6. doi: 10.1002/anie.201302811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gelis I, et al. Structural basis for signal-sequence recognition by the translocase motor SecA as determined by NMR. Cell. 2007;131:756–69. doi: 10.1016/j.cell.2007.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosenzweig R, Kay LE. Bringing dynamic molecular machines into focus by methyl-TROSY NMR. Annu Rev Biochem. 2014;83:291–315. doi: 10.1146/annurev-biochem-060713-035829. [DOI] [PubMed] [Google Scholar]
- 32.Wypijewska A, et al. 7-methylguanosine diphosphate (m(7)GDP) is not hydrolyzed but strongly bound by decapping scavenger (DcpS) enzymes and potently inhibits their activity. Biochemistry. 2012;51:8003–13. doi: 10.1021/bi300781g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sprangers R, Gribun A, Hwang PM, Houry WA, Kay LE. Quantitative NMR spectroscopy of supramolecular complexes: dynamic side pores in ClpP are important for product release. Proc Natl Acad Sci U S A. 2005;102:16678–83. doi: 10.1073/pnas.0507370102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amero C, et al. A systematic mutagenesis-driven strategy for site-resolved NMR studies of supramolecular assemblies. J Biomol NMR. 2011;50:229–36. doi: 10.1007/s10858-011-9513-5. [DOI] [PubMed] [Google Scholar]
- 35.Liu SW, et al. Functional analysis of mRNA scavenger decapping enzymes. RNA. 2004;10:1412–22. doi: 10.1261/rna.7660804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farrow NA, Zhang O, Forman-Kay JD, Kay LE. A heteronuclear correlation experiment for simultaneous determination of 15N longitudinal decay and chemical exchange rates of systems in slow equilibrium. J Biomol NMR. 1994;4:727–34. doi: 10.1007/BF00404280. [DOI] [PubMed] [Google Scholar]
- 37.Hammes GG. Mechanism of Enzyme Catalysis. Nature. 1964;204:342–3. doi: 10.1038/204342a0. [DOI] [PubMed] [Google Scholar]
- 38.Reed MC, Lieb A, Nijhout HF. The biological significance of substrate inhibition: a mechanism with diverse functions. Bioessays. 2010;32:422–9. doi: 10.1002/bies.200900167. [DOI] [PubMed] [Google Scholar]
- 39.Taverniti V, Seraphin B. Elimination of cap structures generated by mRNA decay involves the new scavenger mRNA decapping enzyme Aph1/FHIT together with DcpS. Nucleic Acids Res. 2015;43:482–92. doi: 10.1093/nar/gku1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sinturel F, Brechemier-Baey D, Kiledjian M, Condon C, Benard L. Activation of 5′-3′ exoribonuclease Xrn1 by cofactor Dcs1 is essential for mitochondrial function in yeast. Proc Natl Acad Sci U S A. 2012;109:8264–9. doi: 10.1073/pnas.1120090109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–32. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–67. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.