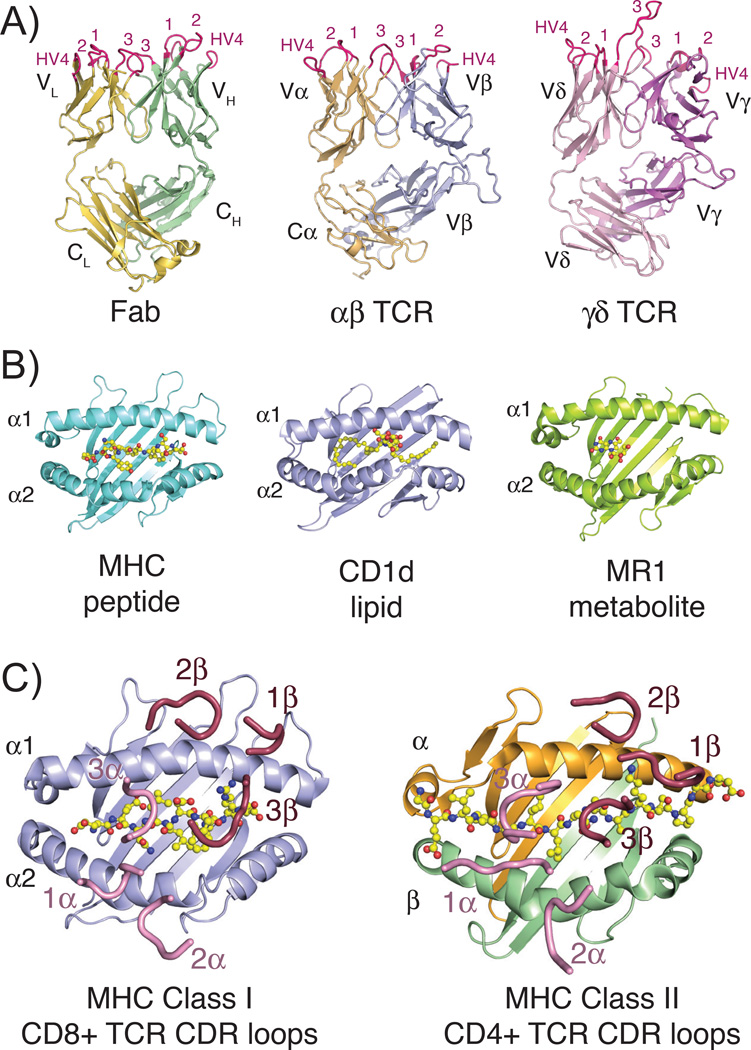
Fig. 1. Structures of TCR, Fab, and MHC molecules and the classical docking paradigm.
(A) Three-dimensional structures of the three classical rearranged receptors in jawed vertebrates: Fab, αβ TCR and γδ TCR (PDB IDs: 1MLC, 2CKB and 1YPZ). Domains are labeled according to their chain designations and CDR loops are colored in hot pink and labeled accordingly. (B) Representative three-dimensional structures of the three antigen-presenting MHC molecules: classical class I MHC with peptide, CD1d with lipid and MR1 with small molecule metabolite (PDB IDs: 2CKB, 1ZT4 and 4LCC). Ligands are shown in stick representations and colored yellow, red and blue indicating carbon, oxygen and nitrogen atoms, respectively. (C) Complex crystal structures of a CD8+ αβ TCR with MHC class I molecule (PDB ID: 3DXA) and CD4+ αβ TCR with MHC class II molecule (PDB ID: 4E41), with just CDR loops shown as they are positioned in the complex structure. Peptide ligands are shown as described above for 1B. In both complexes, CDR loops of the α chain are colored pink, of the β chain are colored raspberry and each are numbered accordingly. All three-dimensional structure figures were made with the program Pymol (Schrödinger).