Figure 2. Whole-exome sequencing of Tet1-deficient tumors reveals mutations of B-non Hodgkin’s lymphoma.
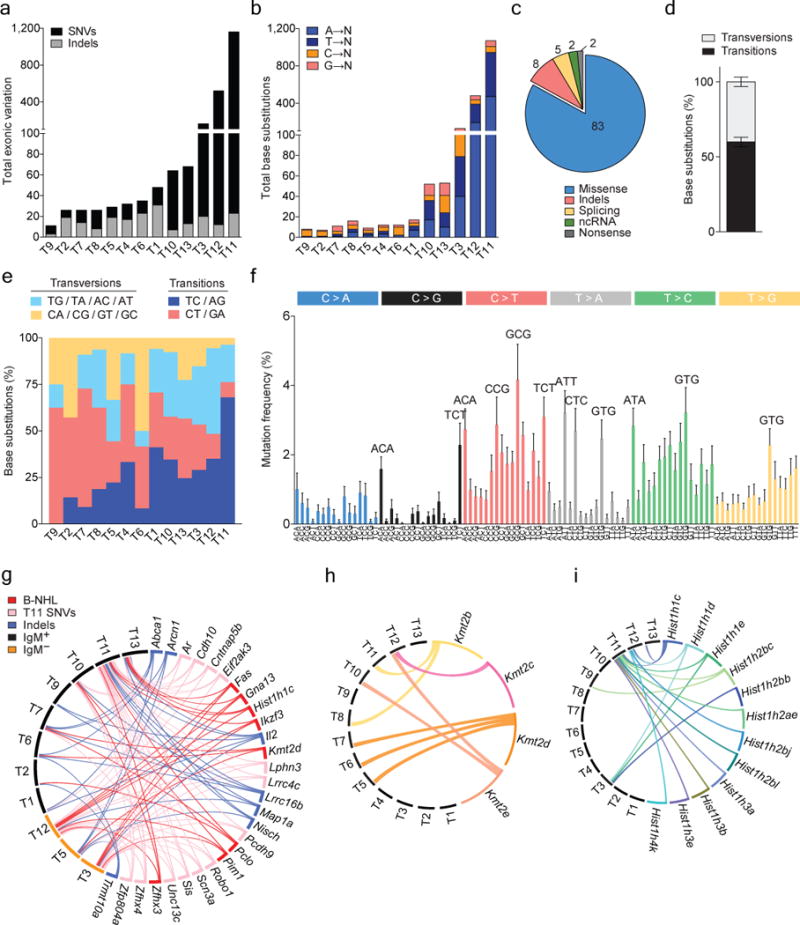
a) Total number of non-synonymous single nucleotide variants (nsSNVs) and insertions and deletions (Indels) detected by whole exome sequencing in thirteen Tet1-deficient tumor cell populations (T1-13) in order of increasing mutation load. b) Total number of nsSNVs divided into A, T, C or G base substitutions, ordered from left to right in tumors according to increasing total exonic variations. c) The frequency of missense, indel, splicing, ncRNA, and nonsense mutations found in Tet1-deficient tumors and d) the average frequency of nsSNVs that are transition and transversion mutations (mean ± SEM, n = 13) and e) according to base substitution frequency per tumor. f) Average mutation frequency of base substitutions in Tet1-deficient tumors according to trinucleotide context (mean ± SEM, n = 13). Circos plots of g) the most frequently mutated genes and their co-occurrence in Tet1-deficient tumors; red = B-NHL recurrently mutated genes, pink = T11 nsSNVs, blue = indels, black = IgM+ tumors, orange = IgM− tumors. Circos plots of mutations and their co-occurrence in h) histone-modifying enzymes and i) histone cluster 1 genes.
