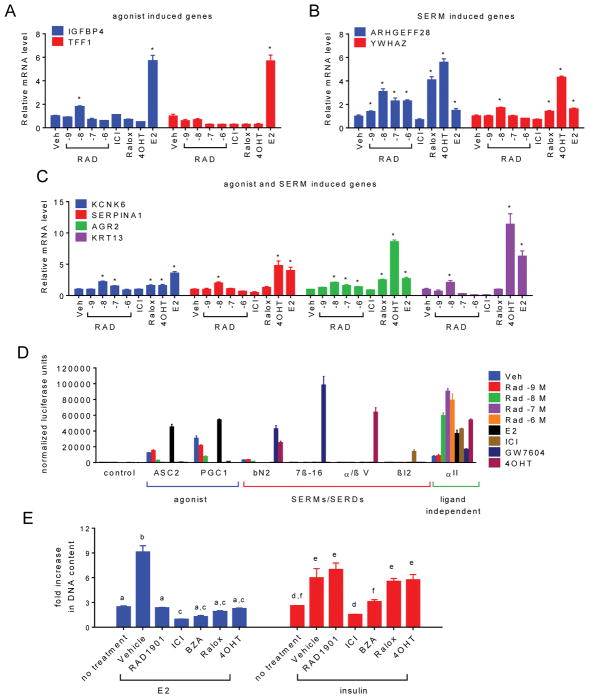
Fig. 5. RAD1901 exerts biphasic agonist/antagonist activity on ESR1 in a dose dependent manner.
A) MCF7 cells were treated for 24 hours with 10−7 M vehicle (Veh), ICI, 4OHT, raloxifene (Ralox), E2 (10−9 M) or RAD1901 (10−9–10−6 M). The expression of ESR1 target genes responsive to (A) agonists, (B) primarily SERMs, or (C) SERMs and agonists was analyzed as in Fig. 1. Relative changes of these and additional target genes designed to evaluate dose dependent response to RAD1901 are presented in Supplementary figure 2. Significant target gene regulation (* p < 0.05) as compared to the vehicle control was detected by 2 way ANOVA followed by Fisher’s LSD test. D) Interaction between ESR1 and conformation-specific peptides in a mammalian two-hybrid system. Triplicate wells of SKBR3 cells were transfected with plasmids expressing ESR1 fused to VP16 together with Gal4DBD alone (control) or Gal4DBD fused to ESR1 interacting peptides noted on the horizontal axis. Cells were then treated with the indicated ESR1 ligands (10−7 M unless otherwise indicated). Interaction of ESR1 with the Gal4DBD peptide constructs was detected through activation of a Gal4-responsive luciferase reporter construct and was normalized to detected β-galactosidase activity expressed in a constitutive manner using a second vector. Normalized response is expressed as fold increase over the detected level of interaction between Gal4DBD alone and ESR1-VP16 in the absence of ligand (Veh). E) The effect of SERMs and SERDs (10−6 M) on the proliferation of MCF7 cells in response to E2 (10−9 M) or insulin (2 × 10−9 M) was evaluated as in Fig. 1. Statistically similar treatments (2-way ANOVA followed by Bonferroni multiple comparison) are indicated by letters.