Figure 1.
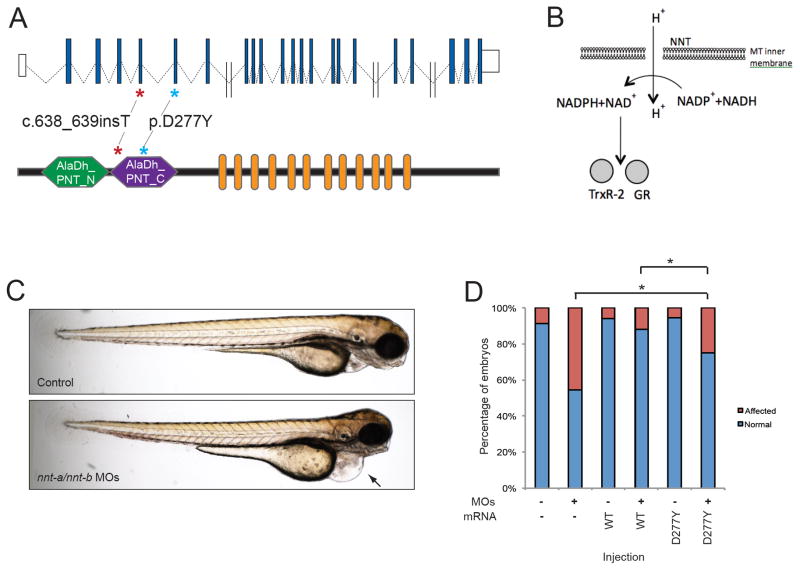
Genetic and Functional evaluation of NNT in LVNC. A. Schematic of the human NNT locus on human chromosome 5 (top) showing non-coding exons (white boxes), and coding exons (blue boxes). Double slash indicates large introns (> 5kb). A schematic of NNT protein is shown below with functional domains Alanine dehydrogenase/PNT, N-terminal domain (AlaDh_PNT_N, shown in green), Alanine dehydrogenase/PNT, C-terminal domain (AlaDh_PNT_N, shown in purple) and twelve transmembrane domains (orange). The two mutations identified in LVNC families are shown with asterisks. B. Schematic of NNT function in mitochondria to regenerate NADPH in the cell; NADPH is then used to regenerate Thioredoxin Reductase 2 (TrxR-2) and glutathione reductase (GR). C. Lateral view of representative 3 dpf control (top) and nnt-a/b morphant (bottom) larvae showing cardiac edema (arrow). D. In vivo complementation assay of NNT p.D277Y; quantification of cardiac edema in 3 dpf morphant zebrafish larvae (5 ng nnt-a/b MOs) with and without addition of 200 pg human wild type NNT RNA (WT), and mutant RNA (p.D277Y). Statistically significant differences were calculated with χ2 tests and p<0.0001 are indicated (*); n=46–51 embryos/injection batch, repeated three times with masked scoring.