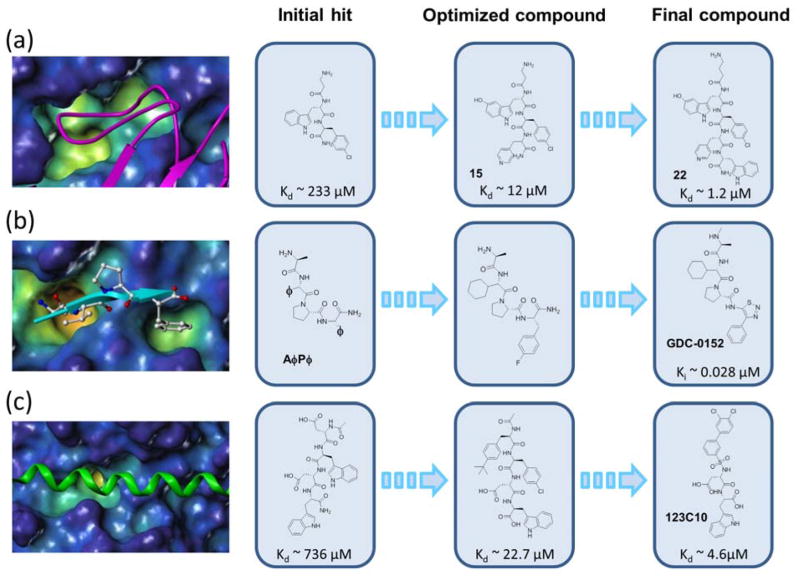
Figure 2. Possible applications of HTS by NMR in targeting PPIs.
a) Example of the discovery of a potent EphA4 inhibitor, targeting a protein-loop interactions. The EphA4 ligand binding domain (surface representation) in complex with ephrin A5 (magenta ribbon) is shown on the left (PDB ID: 4M4R). The evolution into a potent antagonist is illustrated with chemical structures at each step. b) Application of the HTS by NMR to protein-β-strand interactions. The surface representing the BIR3 domain of XIAP in complex with peptide AVPF (cyan ribbon) is shown on the left. (PDB ID: 2OPZ). The HTS by NMR identified the correct essential known consensus AφPφ signature of the peptide. In a second screen using a POS library made up by non-natural aminoacids, a consensus having in particular a cyclohexyl-Glycine in P2 and a 4-F-Phenylalanine in P4 was identified. This consensus motif closely resembles the molecule derived by Genentech, GDC-0152. c) Example of the discovery of compound 121C10 as an inhibitor of Mcl-1, representing an antagonist of protein-α-helix interactions. The surface representing Mcl-1 in complex with Bim BH3 peptide (green ribbon) is shown on the left (PDB ID: 2NL9). The evolution of 121C10 from the initial hits is shown on the right, where the chemical structures and the dissociation constant values (Kd) of selective compound at each step are reported.