Figure 4. Correlation of residue accuracy with confidence values based on the consensus between FR-TM-align, FATCAT, MATT and DaliLite alignments.
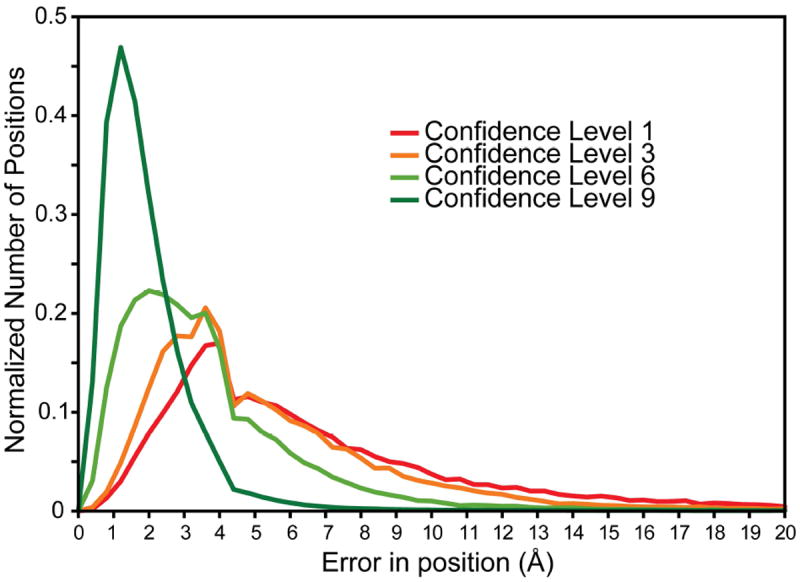
From all consensus alignments of the α-helical subset of HOMEP3, the position-specific confidence level was extracted for positions in which amino acids were aligned, i.e., excluding gapped positions. For each considered position, the distance (in Å) of the corresponding Cα-atom in the homology model to that in the native X-ray structure was calculated. This value then was averaged over the models built based on each of the alignments from the four structural alignment methods. The plot contains the normalized distribution of averaged Cα distances for each of the confidence levels (see Fig. 3).
