Fig. 6.
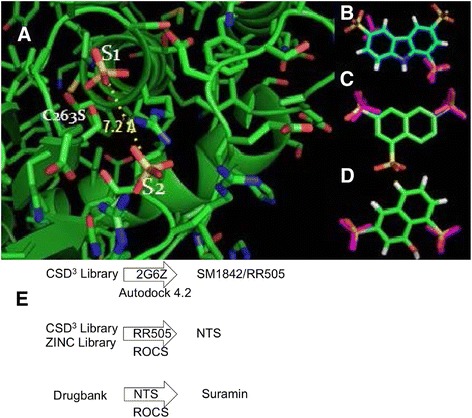
Pharmacophore-based Identification of DUSP5 PD Inhibitors. a Crystal structure of DUSP5 PD(C263S) [16], showing the two bound sulfate ions in the two anion-binding pockets postulated to be occupied by the two phosphate groups of the ERK2 activation loop (pThr-Glu-pTyr) [16–18]. The anion pocket closest to the catalytic nucleophile (Cys263) is labeled S1, and the distal anion pocket is labeled S2. The S2 anion (sulfate) is stabilized be several arginine residues, while the S1 anion may derive some helix dipole stabilization by virtue of its location at the N-terminal end of a long central helix. The sulfur to sulfur distance of 7.2 Å defines the DUSP PD pharmacophore as two anionic groups separated by ~7 Å. Overlay of the S1-S2 pharmacophore (two sulfates, shown as purple) on RR505 indicates a poor match, while (b) overlay on NTS (c) in one of two possible orientations (related by a 180° rotation) is better. d A ligand-based search using this pharmacophore identified CSD 3 _2320, which also matched the S1-S2 sulfate positions well. The overlay in panel (d), as in panel (c), is shown in one of the two possible orientations that optimally align active site sulfate and ligand sulfonate groups. e Flow chart summarizing the docking and ROCS alignment procedures used to identify lead molecules. Once SM1842/RR505 was identified from the CSD3 Library, it was used as a ROCS query and searched against the CSD3 Library and ZINC Library. NTS was identified from the ROCS search. NTS was used as a ROCS query to search Drugbank, which led to identification of Suramin
