Fig. 7.
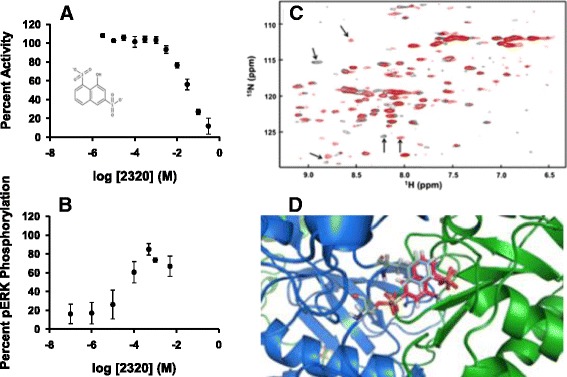
CSD 3 _2320 binding to DUSP5 PD. a Dose response curve for CSD 3 _2320 as an inhibitor of the DUSP5 PD(WT) phosphatase activity, using pNPP as substrate. Experimental conditions as in Figs. 3 and 4. Chemical structure of CSD 3 _2320 in the insert. b Dose response curve for CSD 3 _2320 as an inhibitor of the DUSP5 (full-length protein) phosphatase activity, using pERK2 as a substrate. c DUSP5 PD(C263S) 1H-15N HSQC spectrum of DUSP5 PD( C263S) in pH 6.8, 50 mM potassium phosphate, 100 mM potassium chloride buffer. Overlay is of 500 μM 15 N-labeled DUSP5 PD alone (black), and in the presence of 500 μM CSD 3 _2320 (red). Potentially important chemical shift perturbations due to binding are indicated using arrows. d The model from Fig. 1, with CSD 3 _2320 positioned such that its two sulfonate groups are optimally overlaid with the two phosphate groups on the ERK2 pThr-Glu-pTyr peptide. This overlay results in the phenolic ring of the CSD 3 _2320 naphthalene core being superimposed directly on the tyrosine phenol ring of the pThr-Glu-pTyr peptide
