Fig. 4.
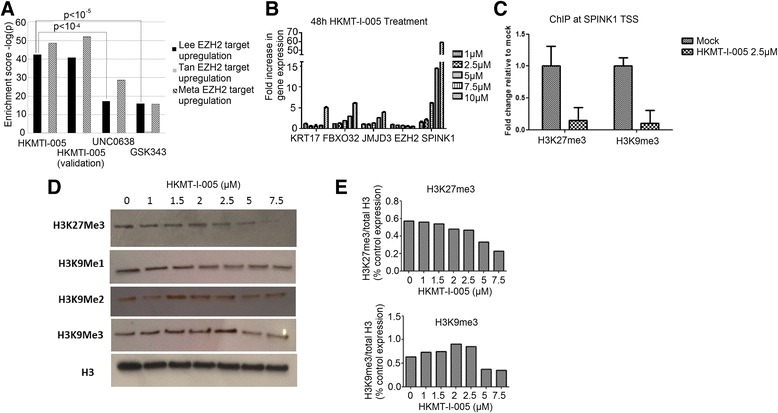
Compound-induced upregulation of EZH2-repressed target genes. a Enrichment scores for differential expression of EZH2 targets on treatment with panel of compounds. Enrichment scores are negative logarithm of p values, such that higher values indicate more significant enrichment. Left-hand bars show enrichment of targets derived from siRNA knockdown of EZH2 in MDA-MB-231 cell line (labelled ‘Lee EZH2 target upregulation’), middle bars show enrichment of targets derived from siRNA knockdown of EZH2 in MCF7 cell line (labelled ‘Tan EZH2 target upregulation’) and right-hand bars show enrichment of targets defined by meta-analysis of 18 independent microarray studies profiling effects of shRNA-mediated EZH2 knockdown in a variety of cell lines (labelled ‘Meta EZH2 target upregulation’): See ‘Materials and Methods’. b Sybr green real-time PCR mRNA level measurement of EZH2 target genes and executing enzymes following a 48-h treatment with HKMTI-1-005 at different concentrations of MDA-MB-231 cells. c Sybr green real-time PCR measurement of the SPINK1 transcription start site following chromatin immunoprecipitation, using antibodies to the histone marks shown, of MDA-MB-231 cells treated with HKMT-I-005 or HKMT-I-011 at 2.5 μM for 24 h. Each IP-value has been determined as the relative increase to the no-antibody control and is shown as fold difference relative to the untreated control. d Western blot showing levels of modified histones, following a 48-h treatment with HKMTI-1-005 at different doses. Total H3 levels are shown for comparison. e Densitometry quantification of Western blot intensity, showing ratio of modified (H3K27me3 top, H3K9me3 bottom) H3 relative to total H3 with increasing dose of HKMTI-1-005 treatment
