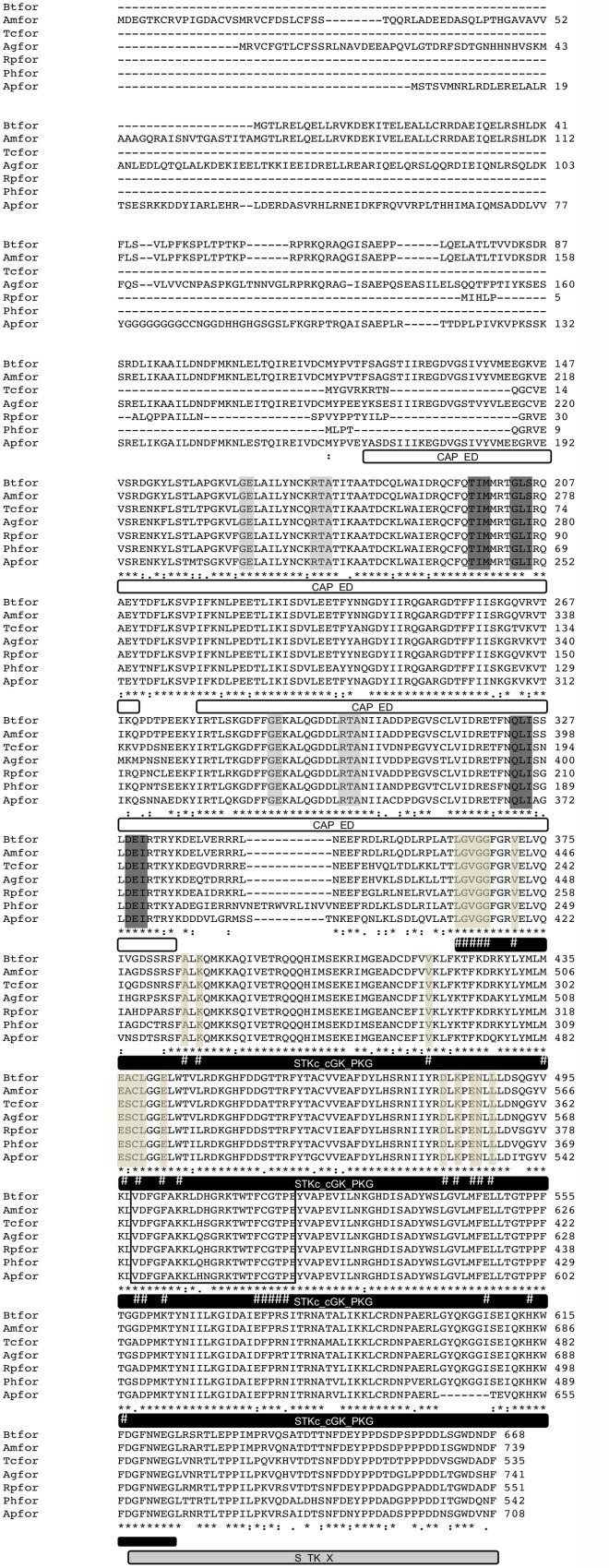
Fig 2. Amino acid sequence of R. prolixus foraging (Rpfor) protein.
The putative coding sequence of the R. prolixus foraging gene is shown in comparison with orthologues from other insect species showing a high degree of conservation. Asterisks indicate identical amino acids, double points represent conserved exchanges and single points indicate homologous amino acids. The predicted protein for the Rpfor gene (code: RPRC000321-PA) contains 551 amino acids (1,656 bp) and a total of fourteen exons. The sequenced amplicon (from the position 195 to 1597) was identical to the Rpfor predicted protein sequence. The four conserved domains identified in the Rpfor gene are shown: two CAP-ED domains (effector domain of the CAP family of transcription factors), one STKc_cGK_PKG domain (catalytic domain of the cGMP- dependent protein kinase) and one S_TK_X domain (extension to Ser/Thr-type protein kinases). The second CAP-ED domain, the ATP binding sites and the active sites of the PKG catalytic domain are conserved between the species used in our analyses. Interestingly, the activation loop of the PKG catalytic domain presents three variable amino acid positions (378, 380 and 381). The first one presents a conserved amino acid change between arginine and lysine, both with polar and positively charged side chains. In the second position, aspartic acid, histidine and glutamine are present. The three amino acids are polar and present charged side chains: positive for histidine and glutamine, and negative for aspartic acid. Finally, the third position is the most variable considering amino acids properties and includes the polar amino acids: asparagine, histidine and serine. The light gray and the dark gray boxes indicate the ligand binding sites and the flexible hinges of the CAP-ED domains, respectively. The light brown boxes indicate the active sites of the STKc_cGK_PKG domain. The ATP binding sites are marked with #. The black box indicates the activation loop region. Sequence features were annotated manually using the Rpfor sequence as a reference. The Rpfor protein sequence has a similar length to those from T. castaneum and P. humanus with 535 and 542 amino acids, respectively. B. terrestris, A. mellifera, An. glabripennis and Ac. pisum present longer N-terminal regions (S1 Text).