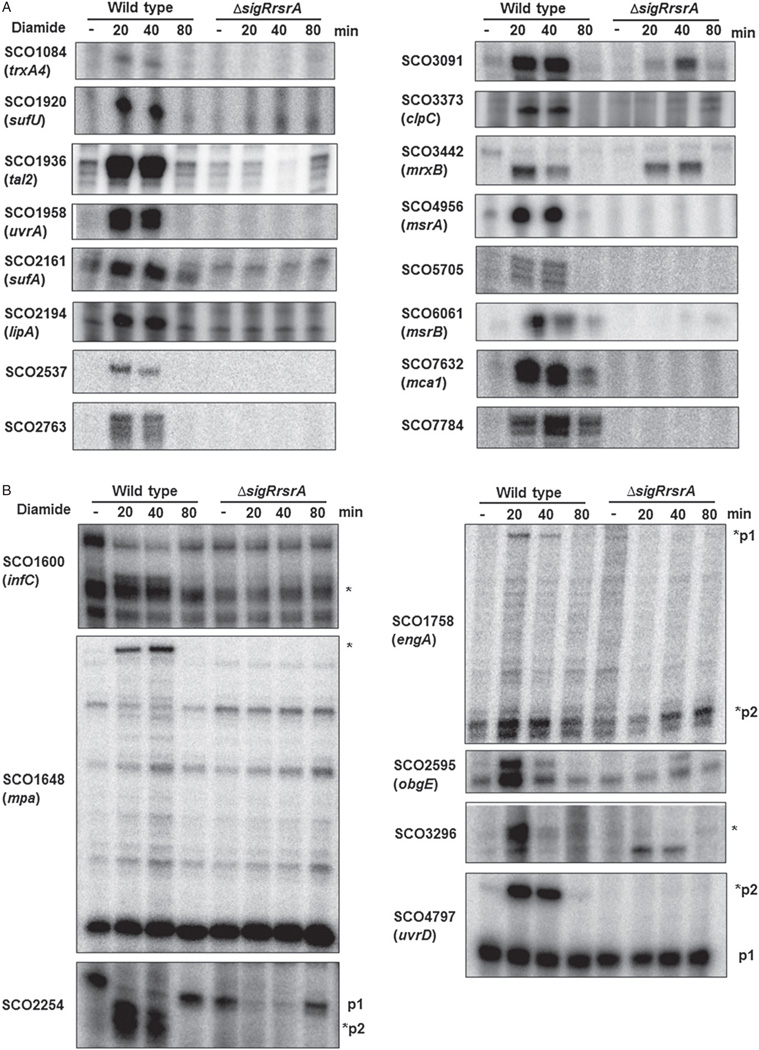
Fig. 1.
S1 nuclease mapping of selected SigR-dependent promoter regions. RNA samples were prepared from exponentially growing wild-type and ΔsigRrsrA cells treated with 0.5 mM diamide for 0, 20, 40 and 80 min.
A. Transcript patterns of genes whose primary transcripts are from SigR-dependent promoters.
B. Results from genes which produce transcripts from other promoters independent of SigR. The bands with marked asterisks in panel B are from predicted SigR-binding promoters.