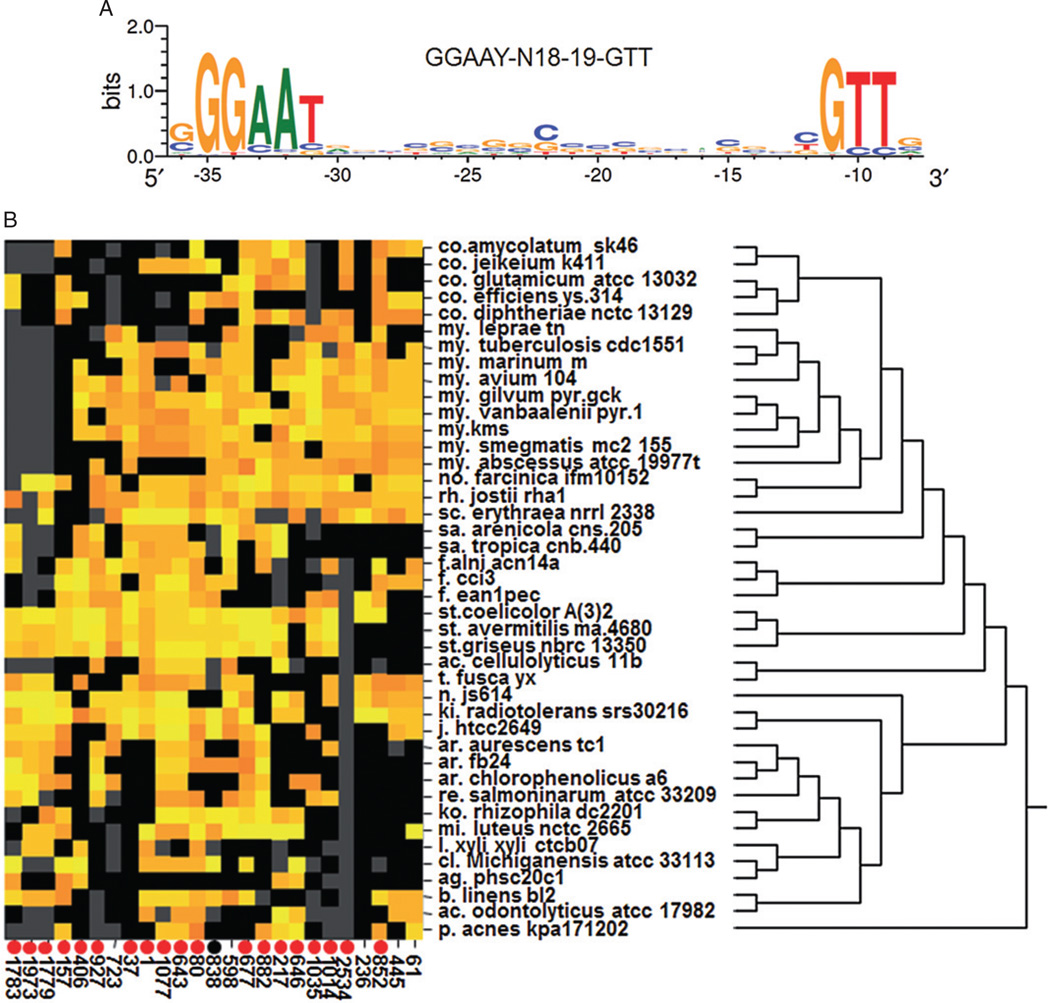
Fig. 4.
The top 25 orthologous sets of genes predicted to be SigR targets across 42 selected Actinomycete species from 22 genera.
A. Sequence logo of the SigR promoter motif in S. coelicolor. The logo was derived from the alignment of 58 SigR-dependent promoters detected by ChIP-chip. The heights of the letters represent their conservation at the particular position of the multiple sequence alignment. The −35 and −10 regions are separated by a non-conserved spacer region of 18 or 19 base pair. The logo was produced using WebLogo (http://weblogo.berkeley.edu/).
B. The heatmap indicates whether the promoter region of the corresponding gene contains a putative SigR promoter for each species (row) and each set of orthologous genes (column). Orange and yellow indicate moderate and strong match to the SigR promoter model respectively. Black indicates that the corresponding species possess a gene belonging to the corresponding orthologous set but that no match to the SigR promoter model, while grey indicates that the species lacks a gene representing the orthologous set. Species are organized according to their phylogenetic relationship as indicated by the dendrogram on the right. Sets of orthologous genes are identified by arbitrary numbers (Table 2). Sets of orthologous genes containing confirmed SigR target genes in S. coelicolor are indicated by circles at the bottom. Red circles indicate genes detected by SigR ChIP-chip and experimentally verified for SigR-dependent expression. The black circle indicates a gene that is associated with a SigR ChIP-chip peak without experimental verification.