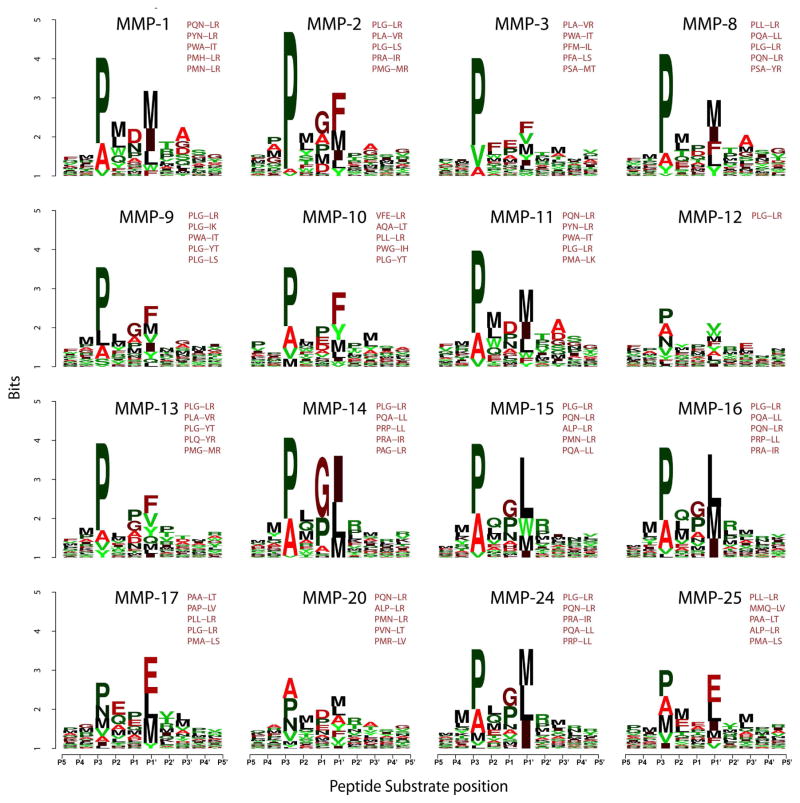
Fig. 4. Frequency plot of the efficient cleavage sequences of MMPs in an IceLogo format.
The height of a character is proportional to the frequency of the amino acid residue at the individual position of the cleaved peptide. 18,583 peptide substrates were cleaved by the individual MMPs. The scissile bond is between the P1 and P1′ residues. The Z-scores for the substrates were calculated and the substrates were ranked according to their Z-scores. The most efficient 100 substrates (Z-score >2; p-value <0.02) were selected for each MMP. Because the design of the peptide substrates was biased to the PXXL-containing sequences, the resulting position-specific matrix of the top 100 substrates was normalized at each position by using the amino acid residue frequency at this particular position in the entire 18,583 peptide library. The five most frequently occurring sequence motifs for each MMP are also shown.